Arginine methylation in global control of RNA splicing and translation
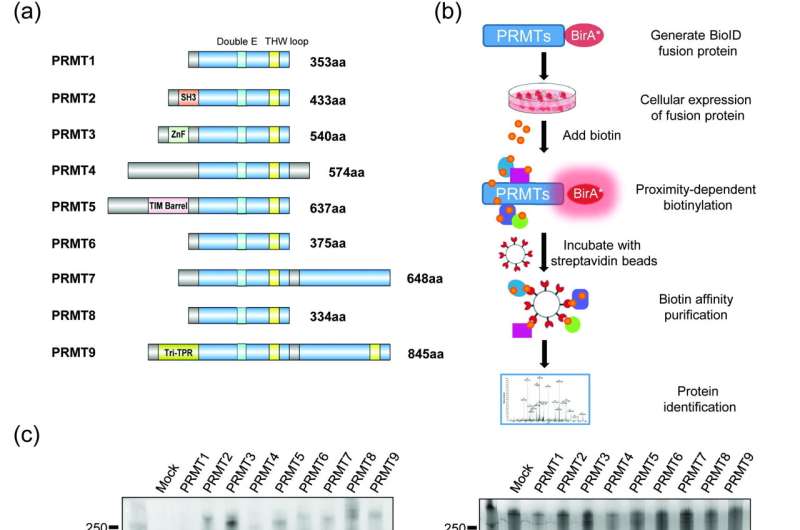
Thousands of proteins undergo arginine N-methylation, a widespread post-translational modification catalyzed by several protein arginine methyltransferases (PRMTs) that covalently link methyl groups to arginine side chains. Nine PRMTs, PRMT1 to PRMT9, have been identified in the human genome, which are further classified into three types according to the final methylarginine products. Arginine methylation plays key roles in various cellular processes.
However, global understanding of their molecular mechanisms and biological functions is limited due to the lack of a complete picture of the catalytic network for each PRMT as well as the absence of reliable antibodies and mechanically well-defined "erasers" and "readers".
In a study published in Science Bulletin, the researchers from the Shanghai Institute of Nutrition and Health (SINH) of the Chinese Academy of Sciences (CAS) identified the interacting proteins for all human PRMTs, and demonstrated their functional importance in mRNA splicing and translation.
Using a sensitive BioID technology, the researchers systematically characterized the interactome and the substrate specificity of all known human PRMTs. They demonstrated significant overlapping of interactomes of human PRMTs with the known methylarginine-containing proteins. Different PRMTs are functionally redundant with a high degree of overlap in their substrates and high similarities between their putative methylation motifs.
Through bioinformatics analysis and experimental validation, the researchers revealed that RNA-binding proteins (RBPs) involved in regulating RNA splicing and translation are highly enriched in PRMT interactomes and undergo extensive arginine methylation, indicating their importance in regulating RNA metabolism.
By applying RNA-seq and Ribo-seq, they demonstrated that the inhibition of PRMTs leads to global alteration of splicing and translation, providing a genome-wide picture of the general functions of protein arginine methylation. Interestingly, they found that the inhibition of PRMT activity did not change the distribution of ribosome occupancy in different regions of mRNAs, suggesting that PRMTs affected the maturation of ribosomes before they are assembled onto the mRNAs.
Using RPS2 as an example, the researchers found that mutations in RPS2's methylation sites suppress ribosome assembly, translation, and eventually cell growth, indicating that arginine methylation of ribosomal proteins is critical to ribosomal assembly. This might be the main mechanistic reason for why PRMT affect translation.
This study provides new insights into biological functions of PRMTs and links individual PRMTs to their arginine methylation events, revealing critical functions of arginine methylation in regulating RNA splicing and translation.
More information: Huan-Huan Wei et al, A systematic survey of PRMT interactomes reveals the key roles of arginine methylation in the global control of RNA splicing and translation, Science Bulletin (2021). DOI: 10.1016/j.scib.2021.01.004
Provided by Chinese Academy of Sciences