Researchers discover a new diet-dependent mechanism for regulating RNA maturation
Particularly sensitive to chemical modifications, messenger RNAs (mRNAs) are molecules responsible for transmitting the information encoded in our genome, allowing for the synthesis of proteins, which are necessary for the functioning of our cells. Two teams from the University of Geneva (UNIGE), Switzerland, in collaboration with the Norwegian University of Science and Technology (NTNU), have focused on a specific type of chemical modification—called methylation—of mRNA molecules in the small worm Caenorhabditis elegans. They found that methylation on a particular sequence of an mRNA leads to its degradation and that this control mechanism depends on the worm's diet. These findings are published in the journal Cell.
Several steps take place before a DNA-encoded gene produces the corresponding protein. One of the two strands of DNA is first transcribed into RNA, which then undergoes several processes, including splicing, before being translated into a protein. This process removes unnecessary non-coding sequences (introns) from the gene, leaving only the protein-coding sequences (exons). This mature form of RNA is called messenger RNA (mRNA).
A "Post-It" to block protein synthesis
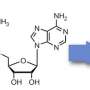
In addition to these processes, RNA—but also DNA molecules—can undergo a chemical modification: methylation. This consists of adding a methyl group (CH3), which modifies the fate of these molecules without altering their sequence. Deposited on the RNA or DNA in very specific places like Post-Its, methyl groups indicate to the cell that a particular fate must be given to these molecules. Methylation of RNA is essential: mice without RNA methylation die at an early embryonic stage.
Two neighboring teams at the UNIGE, one working on RNA regulation and the other specializing in DNA organization in the worm C. elegans, have studied the role of methylation in controlling gene expression. The laboratories of Ramesh Pillai and Florian Steiner, professors in the Department of Molecular Biology at the UNIGE Faculty of Science, have shown for the first time that methylation at the end of the intron of a particular gene blocks the splicing machinery. The intron cannot be removed and the protein is not produced.
Fine regulation to ensure a fair balance
This gene, whose mRNA is modified by methylation, encodes for the enzyme that produces the methyl donor. "It is therefore a self-regulating mechanism since the gene involved in producing a key factor required for methylation is itself regulated by methylation," explains Mateusz Mendel, a researcher in the Department of Molecular Biology at the UNIGE Faculty of Science, and the first author of this study.
Moreover, this modification is dependent on the quantity of nutrients received by the worms. "When nutrients are abundant, the mRNA is methylated, gene splicing is blocked, and the level of methyl donors decreases, which limits the number of possible methylation reactions. On the other hand, when there are few nutrients, there is no methylation of the particular RNA of this gene, so splicing is not blocked and the synthesis of methyl donors increases," says Kamila Delaney, a researcher in the Department of Molecular Biology at the UNIGE Faculty of Science. Elements present in the food provide the raw materials required for producing the methyl donor, so methylation-dependent splicing inhibition puts a brake on its production under conditions of a rich diet. "Aberrant methylation reactions—too much or too little—are the cause of many diseases. The cell has set up this very sophisticated regulatory system to ensure a fair balance of methylations in the cell," says Mateusz Mendel.
Methylation of mRNAs at these specific sequences was discovered in the 1970s by scientists, including Ueli Schibler, a former professor at the UNIGE, before being forgotten. It took 40 years before researchers rediscovered its importance in gene regulation in 2012. With this study, scientists from the Department of Molecular Biology highlight the crucial role of methylation in the control of splicing and in the response to environmental changes.
More information: Cell (2021). DOI: 10.1016/j.cell.2021.03.062
Journal information: Cell
Provided by University of Geneva