This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Researchers develop new method for peptide sequencing based on nanopore sensing technology
New protein sequencing technology with improved sensitivity and throughput will bring revolution to proteomics and clinical diagnostics.
In a study published in Nature Methods, a research team led by Prof. Wu Haichen from the Institute of Chemistry of the Chinese Academy of Sciences (CAS), and Prof. Liu Lei from the Institute of High Energy Physics of CAS, together with their collaborators, have developed a new method for peptide sequencing based on host-guest interaction-assisted nanopore sensing.
The history of protein sequencing could be dated back to the determination of the complete amino acid sequence of insulin by Sanger in the 1950s. However, so far, there are only two major methods for protein sequencing, i.e., mass spectrometry and Edman degradation.
During the past few decades, nanopore sensing emerged as the latest "disruptive" single-molecule technique and has gained great success in the new generation of DNA sequencing development. This has inspired scientists to transplant the technology to single-molecule protein sequencing. However, nanopore sequencing of proteins faces tremendous challenges, such as the realization of a unidirectional transport of heterogeneously charged peptide chains through a nanopore, and electrical identification of 20 individual amino acids or their combinations.
In this study, the researchers proposed an alternative sequencing strategy based on an improved host-guest interaction-assisted nanopore sensing technique.
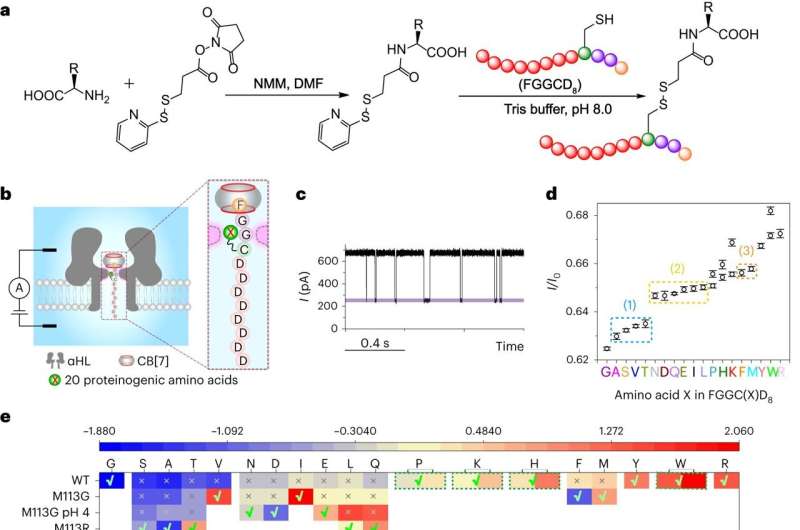
The model peptides were first digested with carboxypeptidases to afford a mixture of amino acids. The next key step was to couple the released amino acids to a FGGCD8⊂CB[7] peptide probe via a covalent linker and then subject the complex to translocation experiments through wildtype α-hemolysin or its mutants.
Finally, the current blockage of each FGGC(X)D8⊂CB[7] peptide was used to identify the amino acid X and their relative abundance was used to determine the order of the enzymatic cleavage, i.e., the sequence of the peptide.
This study serves as a proof-of-concept demonstration for a novel method capable of precisely determining the amino acid sequence of a peptide. While notable limitations persist, this marks a significant advancement and unveils a promising avenue for the future of protein sequencing.
More information: Yun Zhang et al, Peptide sequencing based on host–guest interaction-assisted nanopore sensing, Nature Methods (2023). DOI: 10.1038/s41592-023-02095-4
Journal information: Nature Methods
Provided by Chinese Academy of Sciences