For the first time, DNA and proteins sensed by de novo-designed nanopore
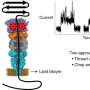
Researchers in Japan have designed the first bottom-up designed peptides, comprising chains of amino acids, that can form artificial nanopores to identify and enable single molecule-sorting of genetic material in a lipid membrane.
Biological nanopores are generally channels made by pore-forming proteins, that can detect specific molecules, but such natural channels are difficult to identify, limiting proposed applications in low-cost, speedy DNA sequencing, small molecule detection and more.
"Nanopore sensing is a powerful tool for label-free, single-molecule detection," said corresponding author Ryuji Kawano, professor in Tokyo University of Agriculture and Technology (TUAT) in Japan. "This is the first time that DNA and polypeptides were sensed using a de novo-designed nanopore."
They published their findings on November 22 in Nature Nanotechnology.
The de novo-designed nanopores are built "from scratch," according to Kawano, and have the potential to mimic natural proteins and their ability to detect specific proteins. Crucially, Kawano said, they can also be engineered to act as artificial molecular machines capable of detecting a much wider range of molecules—which may help elucidate the connection between structure and function in target proteins.
"The folded structure of proteins is determined by their linear polypeptide sequence and gives rise to specific protein functionality," Kawano said, noting that all proteins have a unique structure and size. "The unique primary structure is the result of structural evolution such as the mutation and selection of amino acid residues over time. To reveal the relationship between this primary information and protein structure is one of the ultimate goals of science."
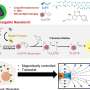
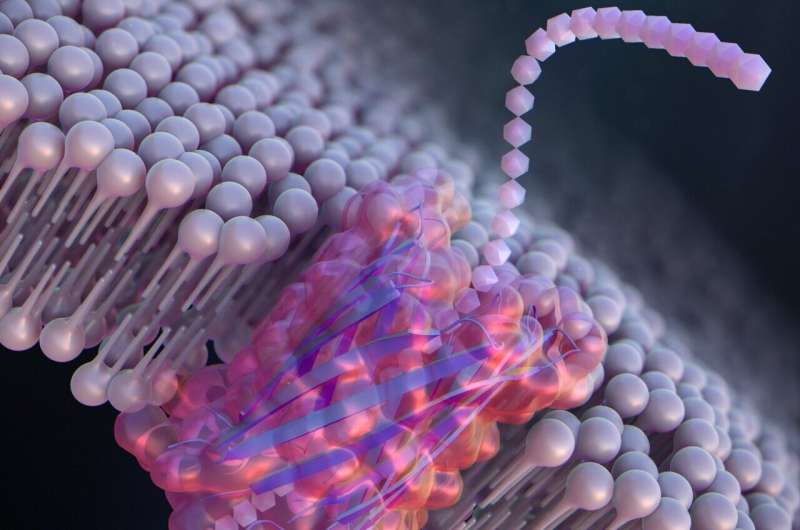
To develop large synthetic nanopores that can better detect and identify molecules for practical applications, Kawano and the team designed a peptide dubbed SV28. With two arms of amino acids bent at a sharp angle, and specific charges at the terminus, the orientation of the hairpin-shaped peptide can be precisely controlled by applying a voltage. The peptide can assemble to form nanopore structures ranging in size from 1.7 to 6.3 nanometers, suitable for detecting molecules of DNA.
The researchers also modified SV28 by adding a mutation that causes the peptide structure to bend and twist in specific ways. The resulting peptide formed evenly dispersed pores of 1.7 nanometers each, capable of detecting a single polypeptide chain—or one half of a protein.
This achievement could be applied to facilitating the understanding of the relationship between protein structure and function.
For the next steps, the team plans to design various peptides and proteins to construct different types of nanopores to aid in peptide sequencing, operate as molecular robots, and more.
More information: Keisuke Shimizu et al, De novo design of a nanopore for single-molecule detection that incorporates a β-hairpin peptide, Nature Nanotechnology (2021). DOI: 10.1038/s41565-021-01008-w
Journal information: Nature Nanotechnology
Provided by Tokyo University of Agriculture and Technology