BridgIT, a new tool for orphan and novel enzyme reactions
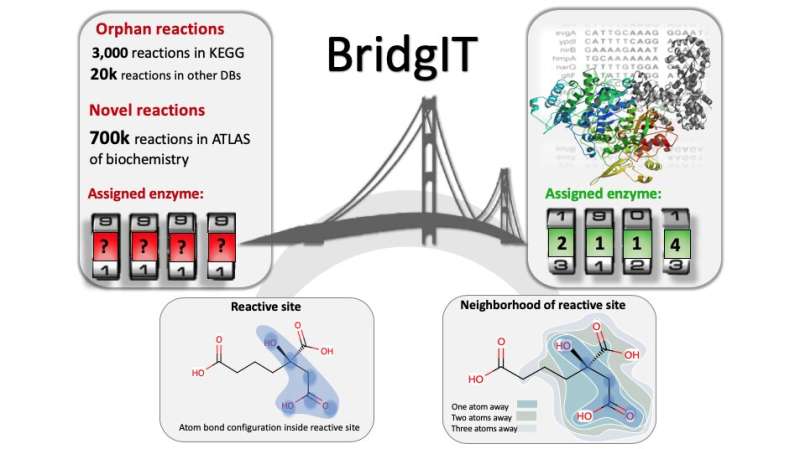
Effective protein engineering can give us control over the generated products inside a cell. However, for many of the biochemical reactions responsible for these products, we don't we don't know the specific protein or enzyme-producing gene responsible. These so-called orphan reactions have become a big problem for protein engineers.
Moreover, software that predicts novel, hypothetical biochemical reactions—a common tool for modern biochemists and synthetic biologists—cannot assign potential genes to them, meaning that there are no recorded DNA sequences that scientists can tweak to change protein or enzyme production. And to further complicate matters, there are also many orphan metabolic enzymes whose particular reaction is unknown, thus leaving important gaps in the maps of metabolic networks and pathways.
In short, finding which gene(s) correspond to the enzyme/protein(s) that catalyze an orphan or novel, hypothetical reaction has grown into a critical issue for applications ranging from biotechnology to medicine.
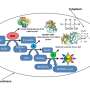
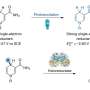
Now, chemical engineers from the lab of Vassily Hatzimanikatis at EPFL have found a solution. The group developed a new computational method and online tool, called BridgIT, to identify candidate genes and catalyzing proteins for hypothetical orphan and novel reactions. All BridgIT needs to know is the four connecting bonds around the atoms of the reactive sites, and it can correctly annotate proteins for 93 percent of analyzed enzymatic reactions. This percentage rose to almost 100 percent when seven connecting bonds were included.
To test BridgIT's accuracy, the researchers pitted it against databases of reactions that were once orphan but have now been assigned to genes and enzymes—basically, reactions that have become non-orphan. BridgIT predicted the exact or a highly related enzyme for 211 out of 234 reactions (>90 percent). And for hypothetical reactions that were once novel and have since been assigned enzymes, BridgIT found the exact enzymes for 334 out of 379 reactions (>88 percent).
The authors write, "BridgIT... will allow researchers to fill the knowledge gaps in metabolic networks and will act as a starting point for designing novel enzymes to catalyze non-natural transformations."
More information: Noushin Hadadi et al, Enzyme annotation for orphan and novel reactions using knowledge of substrate reactive sites, Proceedings of the National Academy of Sciences (2019). DOI: 10.1073/pnas.1818877116
Journal information: Proceedings of the National Academy of Sciences
Provided by Ecole Polytechnique Federale de Lausanne