Directed evolution of a designer enzyme with an unnatural catalytic amino acid
The impressively high conversion rates of natural enzymes partly result from increasing the catalytic activity of a selected few amino acid side chains through precise positioning within the protein binding cavity. Scientists have now demonstrated that such fine-tuning is also possible for "designer" enzymes with unnatural catalytic amino acids. In the journal Angewandte Chemie, they report that laboratory "evolution" of a designer enzyme with an aniline side chain led to variants with significantly higher activity.
The speed and selectivity with which enzymes in nature catalyze conversions are enviable. To catalytically boost unnatural reactions, researchers mimic enzymes with the help of protein frameworks realized by computer-aided protein design. Further optimization is achieved through repetition of a Darwinian cycle: 1) diversification through mutation, 2) identification of improved catalysts, and 3) amplification of the more efficient enzyme variants. This allows for the production of designer enzymes with very high activities.
Researchers led by Clemens Mayer and Gerard Roelfes at the University of Groningen (the Netherlands) have now demonstrated that this type of directed evolution is also a method for improving the efficiency of a novel class of designer enzymes: enzymes that contain an amino acid that is not utilized by nature.
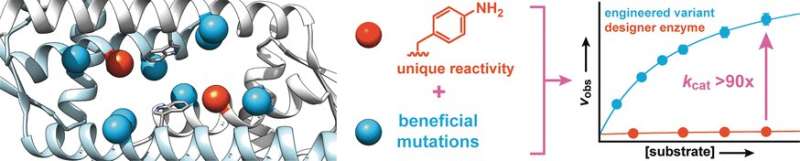
Starting with a protein from Lactococcus lactis, a bacterium used in the production of dairy products such as cheese and buttermilk, the researchers synthesized a designer enzyme that contains an amino acid with an abiotic aniline side chain (aminophenylalanine). Like free aniline, this amino acid catalyzes the reaction of aldehydes with hydrazines or hydroxylamines to make hydrazones or oximes, respectively.
To increase the activity of the enzyme, the researchers produced enzyme variants with mutations at amino acids near the aniline side chain. Screening of about 400 mutants yielded two candidates with better activity, one of which was subjected to a second evolutionary round. This led to the discovery of more beneficial mutations. To identify synergetic effects, multiple favorable mutations were combined to produce further variants. In this way, it was possible to increase the conversion rate of the enzyme by a factor of 90.
The researchers emphasize that, akin to natural enzymes, "this drastic increase is based on strengthening the inherent catalytic activity of the aniline side chain. We intend to use this principle to incorporate further organic catalysts as side chains in enzymes, and to use directed evolution to convert these into highly effective designer enzymes that can rapidly and efficiently carry out synthetically important reactions that would otherwise only run very slowly."
More information: Clemens Mayer et al. Directed Evolution of a Designer Enzyme Featuring an Unnatural Catalytic Amino Acid, Angewandte Chemie International Edition (2018). DOI: 10.1002/anie.201813499
Journal information: Angewandte Chemie International Edition , Angewandte Chemie
Provided by Wiley