This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Research team uncovers universal code driving the formation of all cell membranes
Researchers at the University of Alberta have uncovered what they say has been the missing puzzle piece ever since the genetic code was first cracked.
The code is the universal set of rules that allow living organisms to follow genetic instructions found in DNA and RNA to build proteins. In new research, published in BMC Biology, the U of A team describes a unifying code that guides the binding of those proteins with lipids to form membranes—the wrapper around all cells and cell components.
"Sixty years ago, scientists started to work on how genes encode proteins, but that's not the end of the story," says biochemistry professor Michael Overduin, executive director of the National High Field Nuclear Magnetic Resonance Center. "Along with DNA, RNA and proteins, living cells require membranes. Without the membrane, it's like you've got a house with no walls."
"We theorized that proteins make all membranes rather than membranes just forming magically by themselves, and that hypothesis turned out to be remarkably useful."
'A conceptual revolution'
Overduin says the newly proposed proteolipid code is built from structural insights afforded by new technology and software. The theory describes how membranes are compartmentalized, remodeled and regulated, and provides a basis for understanding fundamental questions such as how life is formed at conception, how viruses invade cells and how neurons send signals for feeling, thought and action.
"We feel that this represents a conceptual revolution that is akin to the discovery of the genetic code," says Overduin. "We're the first to see the overall forest from the trees."
The proposed proteolipid code could also help with drug development for cancer and neurological diseases such as Alzheimer's and Parkinson's that are caused by proteins interacting incorrectly with membranes, Overduin says.
"Using our code, we can now predict how proteins sort in cells and bind lipids, and we can also use polymers to cut out sections of membranes as they are in the heart or the brain and use the resulting disks to perform drug discovery on the real drug targets in the membrane, rather than in an artificial environment with no lipids around," he says.
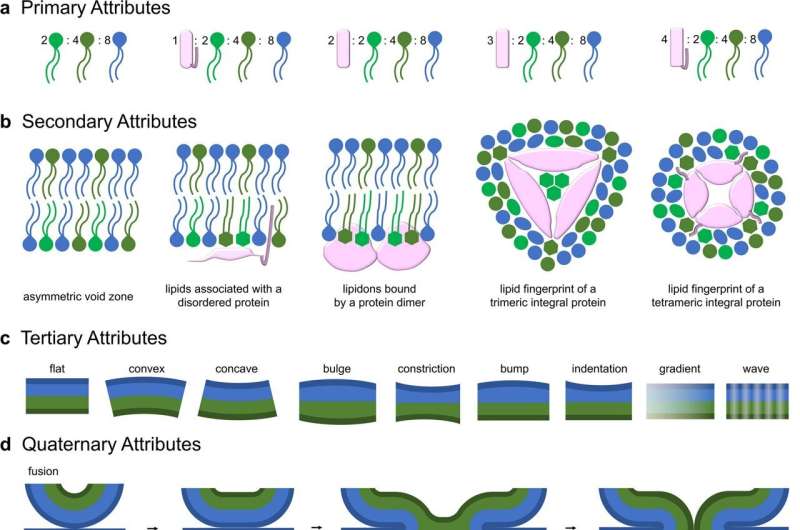
The team suggests four levels of membrane structure, from the simplest to the most complex. Building on the concept of codons—the sequence of three consecutive nucleotides in a DNA or RNA molecule that code for a specific amino acid to make proteins—the team is coining the term "lipidons" to describe the rules for how sets of lipids interact with proteins to form different membranes.
"The lipidon is like a QR code consisting of three lipid molecules that are recognized by proteins," Overduin explains. "It's the proteins that distribute lipids to different places in the cell to give each membrane its unique shape and curvature. Proteins can also make two membranes come together and fuse into one, or they can split a membrane into two different membranes."
Taking a fresh look at an old problem
Overduin admits the proteolipid code turns scientific orthodoxy on its ear and may be hard for some to accept. But he argues that's often how new ideas develop in science, citing several Nobel Prize winners who were originally dismissed for their novel insights.
"I think the difficulty with science is that there often is a herd mentality, and if you want to break the mold and come up with a new approach, there's resistance," he says.
Overduin notes that the first author of the paper, Troy Kervin, was an undergraduate student in his laboratory during the COVID-19 pandemic who is now pursuing a Ph.D. at Oxford University.
"It is remarkable that an undergraduate student can come up with a new code that turns biology upside down and allows us to make sense of it in a radically new way," Overduin says. "It's a nice example of how fresh eyes can take an old problem that has confounded senior scientists for decades and crack it."
"Our undergrads have tremendous energy and enthusiasm, and they come without the biases that other scientists might have," says Overduin, who employs half a dozen undergraduate student researchers in his lab each summer.
Next steps for the research include using the proteolipid code to better understand the specialized membranes of nerve cells, bacteria, viruses and mitochondria, which produce energy for cells.
"We're still in the early days of this fundamental research in terms of translating it into ways to help people," Overduin says.
More information: Troy A. Kervin et al, Membranes are functionalized by a proteolipid code, BMC Biology (2024). DOI: 10.1186/s12915-024-01849-6
Journal information: BMC Biology
Provided by University of Alberta