This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Fluorescent protein outshines the competition when imaging cells
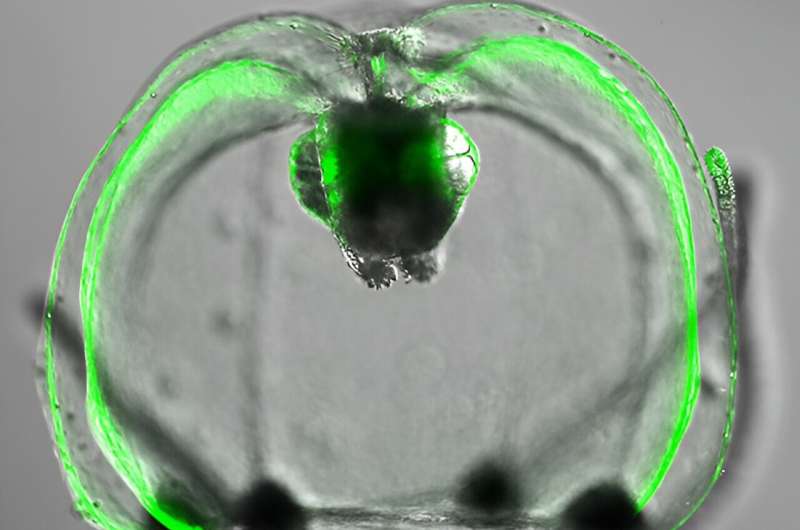
Delicate cellular structures and dynamic processes within cells that were hitherto unseen could be revealed by the next generation of a green fluorescent protein developed by chemists at RIKEN.
Green fluorescent proteins are widely used in biological research for lighting up structures of interest in fluorescence microscopy. They emit green light when illuminated by blue or ultraviolet light.
Previously, Atsushi Miyawaki of the RIKEN Center for Brain Science and his co-workers had derived a green fluorescent protein from a naturally occurring fluorescent protein made by a tiny Japanese jellyfish. It overcame a common problem with green fluorescent proteins, namely they become dimmer under the powerful or long illumination used in fluorescence imaging of live cells.
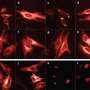
Cellular proteins labeled with the green fluorescent protein, which the team named StayGold, could be tracked within live cells over extended time frames. "It's a highly bright and photostable green fluorescent protein," says Miyawaki. "That makes it useful for imaging a variety of components in cells with an enhanced resolution and over long times."
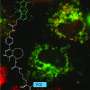
One limitation with the original StayGold, however, was its propensity to pair up and form dimers—molecules made up of two identical building blocks, or monomers. This can cause problems with some proteins.
"The formation of dimers may interfere with the functions of the host proteins to which StayGold fuses," explains Miyawaki. "For example, it may result in cross-linked experimental artifacts in some host proteins."
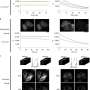
To overcome this limitation, the team determined the green fluorescent protein's crystal structure to identify structural features that may drive the formation of dimers. They then used directed evolution to modify the protein's structure at these key points. Their goal was to suppress dimer formation without impairing the green fluorescent protein's brightness or stability.
Remarkably, the monomer-forming variant that the researchers developed turned out to be even brighter and more photostable than the first generation of StayGold.
The researchers demonstrated the potential of their monomeric variant in several live fluorescence imaging studies. The research is published in the journal Nature Methods.
"We used the monomer to label filaments of actin and the inner membranes of mitochondria," says Miyawaki. "It enabled us to successfully image these structures with an enhanced spatiotemporal resolution and over an extended observation period."
The team is now seeking to elucidate the molecular basis of the outstanding photostability of the variant. They are also striving to develop a photostable, red-emitting fluorescent protein that could be used in combination with StayGold or its monomer for live fluorescence imaging.
More information: Ryoko Ando et al, StayGold variants for molecular fusion and membrane-targeting applications, Nature Methods (2023). DOI: 10.1038/s41592-023-02085-6
Journal information: Nature Biotechnology , Nature Methods
Provided by RIKEN