This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Researchers observe how the flexibility of a protein hinge is crucial to the transfer of cell proteins
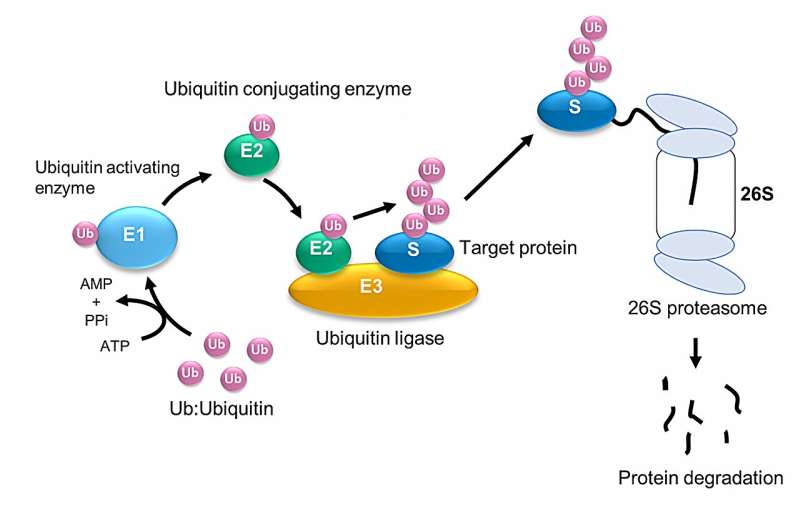
Ubiquitination—the addition of the protein ubiquitin—is a key stage in many cell processes, such as protein degradation, DNA repairs, and signal transduction. Using high-speed atomic force microscopy (HS-AFM) and molecular modeling, researchers led by Hiroki Konno and Holger Flechsig at WPI-NanoLSI, Kanazawa University have identified how the mobility of a ubiquitination-related enzyme hinge allows ubiquitination to take place.
Previous studies have identified a number of enzymes that facilitate ubiquitination, including an enzyme that activates ubiquitin (E1), an enzyme that conjugates it (E2), and an enzyme that catalyzes ubiquitin protein joining (i.e., a ligase, E3) to target protein.
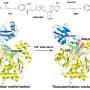
The HECT-type E3 ligase is characterized by a HECT domain that comprises an N lobe with the E2-binding site and a C lobe with a catalytic Cys residue. A flexible hinge connects the two lobes, leading to the hypothesis that ubiquitination is facilitated by the rearrangement of the protein around this hinge.
Konno and their collaborators deployed their high-speed atomic force microscope to hunt for evidence that this was the case. The research is published in the journal Nano Letters.
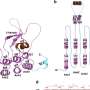
The researchers noted that when the HECT domain was crystallized with a type of E2 enzyme, it formed an L shape such that the distance between the catalytic Cys residue of the HECT domain and the catalytic Cys of the E2 enzyme was 41 Å—too far for the transfer of ubiquitin. However, in its catalytic conformation the HECT domain has a different shape where the distance between the two catalytic Cys residues is much closer—just 8 Å—so this is thought to be a "catalytic conformation."
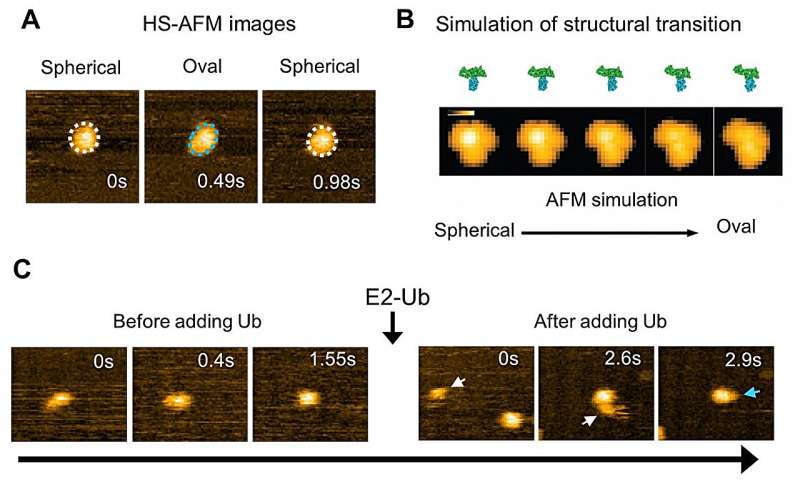
Analysis of HS-AFM images of a wild-type HECT domain of E6AP revealed two conformations—one of which looked spherical and the other oval. Using AFM simulations they attributed the oval shapes to the L conformation and spherical shapes are either the catalytic conformation or the so-called inverted T conformation, which had been observed in another type of HECT domain where the distance between the Cys residues is 16 Å.
To overcome the spatio-temporal resolution limitations of imaging, the experiments were complemented by molecular modeling to visualize HECT domain conformational motions at the atomistic level. Simulation AFM was used to generate a corresponding pseudo AFM movie, which clearly showed the change from spherical to the oval shaped topography.
"Although experimental limitations do not allow us to resolve the intermediate conformations," explain the researchers in their report of the work. "The performed modeling provides evidence that the transitions between spherical and oval HECT domain shapes observed under HS-AFM correspond to functional conformational motions under which the C-lobe rotates relative to the N-lobe, thereby allowing the change between catalytic and L-shape HECT conformations."
Further experiments with mutant HECT domains with less flexibility in the hinge revealed no flipping between conformations—the mutant HECT domains were locked in the catalytic conformation. They also found that these mutant HECT domains could form two ubiquitin proteins joined together more efficiently than the wild type.
E6AP, the HECT-type E3 in this study, interacts with E6 protein derived from human papillomavirus (HPV) and ubiquitinates p53, a tumor suppressor protein. It is also known that ubiquitination of p53 by E6AP and E6 is a major cause of cervical cancer. However, the mechanism of p53 ubiquitination by the interaction of E6AP and E6 proteins remains unclear. In the future, the team will elucidate the structural dynamics of E6AP/E6, and the E6AP/E6/p53 complex with HS-AFM, and clarify how E6 increases the activity of p53 ubiquitination by E6AP.
More information: Kazusa Takeda et al, Structural Dynamics of E6AP E3 Ligase HECT Domain and Involvement of a Flexible Hinge Loop in the Ubiquitin Chain Synthesis Mechanism, Nano Letters (2023). DOI: 10.1021/acs.nanolett.3c04150
Journal information: Nano Letters
Provided by Kanazawa University