Tracking protein disposal could lead to improved therapies

Northwestern Medicine investigators developed a technique to catalog how cells dispose of unnecessary proteins, a process that has implications for cancer and autism-spectrum diseases, according to findings published in Nature Communications.
Defects in ubiquitination, or how cells dispose of unneeded proteins, have been implicated in a variety of diseases, but scientists have previously lacked a reliable method for tracking the enzymes involved in this disposal process, according to Hiroaki Kiyokawa, MD, Ph.D., professor of Pharmacology and co-senior author on the study.
To combat this, a collaborative team led by Kiyokawa and Jun Yin, Ph.D., associate professor of Chemistry at Georgia State University, developed orthogonal ubiquitin transfer, a method of tracking which enzymes trigger disposal of which proteins. This technique may give scientists a better understanding of the mechanisms of disease arising from abnormal ubiquitination, hopefully leading to improved therapy, Kiyokawa said.
Ubiquitination flags proteins for destruction and is facilitated by three classes of enzymes; E1, E2 and E3 enzymes. Ubiquitin is bound to the three enzymes in sequence, but E1 and E2 enzymes are simple primer enzymes, so mammalian cells have just a limited number of E1 and E2 varieties. On the other hand, there are hundreds of E3 enzymes, suggesting each E3 has its own unique protein target.
"The relationship between E3 enzymes and the target protein is like a lock and a key," said Kiyokawa, also a professor of Pathology and a member of the Robert. H. Lurie Comprehensive Cancer Center of Northwestern University.
The diversity of target proteins and weak interactions mean it can be challenging to clearly identify which E3 enzyme tags which protein. However, orthogonal ubiquitin transfer (OUT) is a novel solution to the problem, according to Kiyokawa.
In OUT, scientists engineered an artificial ubiquitin pathway that mimics the native system but only responds to mutated ubiquitin, which has a special tag that allows scientists to identify the protein it eventually targets at the end of the cycle.
"There are two pathways within the same cells," Kiyokawa said. "The original is like a highway, and the engineered pathway is a detour."
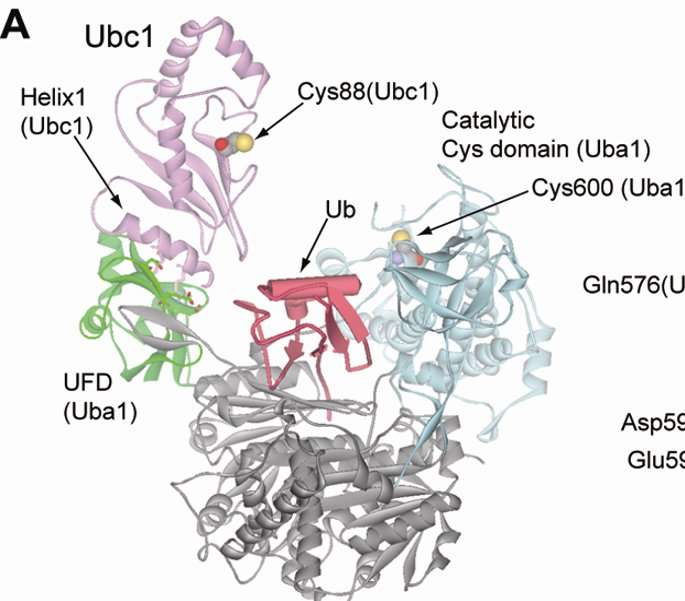
In the current study, the investigators constructed an artificial pathway for an E3 enzyme called E6AP, and introduced the mutated ubiquitin and pathways into cells. After a proteomics screen, the scientists were able to identify the specific protein targets of E6AP.
"This study is just a showcase of the technology, but we believe we can apply this to many of the 600 E3 proteins without too much hassle," Kiyokawa said. "Quite a few E3 enzymes are known to be overexpressed or mutated in neuronal diseases, so there's a large pool of research opportunities."
However, E6AP in particular has been shown to target tumor suppressor proteins when improperly activated by the human papillomavirus, contributing to the increased cancer risk faced by infected patients. In addition, the gene locus that codes for E6AP is known to be mutated in Angelman syndrome, an autism-spectrum disorder.
"We still don't know why E6AP needs to be mutated to show symptoms of Angelman syndrome, but investigation into specific targets may teach us more about the pathophysiological mechanisms of autistic disease," Kiyokawa said.
Further investigation into specific targets is already underway—Kiyokawa and colleagues recently published another study involving OUT, this time in Science Advances.
They found a different E3 enzyme, CHIP, was involved in degradation of CDK4, an enzyme that plays a key role in cell proliferation.
"It seems to be a mechanism that can stop the cell cycle under stress," Kiyokawa said. "If this regulation is broken, the organism may be more susceptible to cancer."
Yet, it's not enough to simply identify these targets of ubiquitination, according to Kiyokawa.
"We need to learn why these newly defined targets are under the regulation of E3 enzymes if we wish to find new therapeutic targets," he said. "Only then can we develop drugs or treatments."
More information: Yiyang Wang et al. Identifying the ubiquitination targets of E6AP by orthogonal ubiquitin transfer, Nature Communications (2017). DOI: 10.1038/s41467-017-01974-7
Karan Bhuripanyo et al. Identifying the substrate proteins of U-box E3s E4B and CHIP by orthogonal ubiquitin transfer, Science Advances (2018). DOI: 10.1126/sciadv.1701393
Journal information: Science Advances , Nature Communications
Provided by Northwestern University