This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Unlocking citrus tolerance secrets: The Valencia sweet orange genome and the fight against HLB disease
Sweet orange (Citrus sinensis L.), a hybrid fruit crop originating from mandarins (Citrus reticulata Blanco) and pummelos [Citrus maxima (Burm.) Merr.], exhibits complex genomic diversity due to ancient interspecific hybridization. At present, the best assembly of the sweet orange genome is the double haploid sweet orange (HSO).
Sequencing of the diploid sweet orange genome only yielded poor quality haploid-sized assemblies, which did not provide a reference level of the genome. The uniformity of sweet orange cultivars makes them prone to diseases like Huanglongbing (HLB), challenging citrus production globally. Therefore, there is an urgent need to better assemble the genome at the chromosome level of sweet oranges and explore genes for HLB tolerance, which is crucial for the sustainable planting and future of sweet oranges.
In November 2022, Horticulture Research published a perspective titled by "A chromosome-level phased genome enabling allele-level studies in sweet orange: a case study on citrus Huanglongbing tolerance."
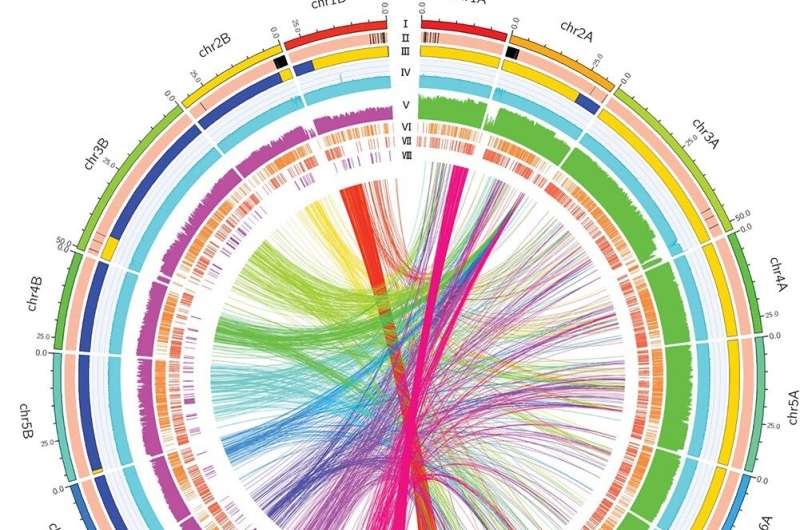
In order to assemble a new sweet orange genome, researchers employed PacBio continuous long reads to decode the DVS genome and obtained a 607.6 Mb assembly with an N50 length of 15.4 Mb using CANU. A phased assembly approach was then applied, leaving 3.9 Mb of homozygosity unphased.
The final assembly connected contigs into 18 pseudo-chromosomes of 598.6 Mb, which were categorized into DVS_A and DVS_B chromosome sets, revealing homologous regions originating predominantly from mandarin and pummelo, respectively. The assembly's error rate was significantly lower than the previously established HSO. Chromosomal comparison of DVS_A and DVS_B to pummelo and mandarins showed 97.1% mandarin-origin regions in DVS_A and 87.4% pummelo-origin regions in DVS_B.
Nucleotide similarity between DVS_A and DVS_B was 96.2%, with numerous variations identified. DVS's gene annotation indicated 55,745 protein-encoding genes and the highest BUSCO completeness among citrus genomes. Allelic expression patterns (AEPs) were studied using RNA-seq data, and DVS served as a superior reference, with significant mapping rates. Various factors, including tissue type, developmental stage, and disease, notably altered AEPs, underlining the complexity of gene expression in sweet oranges.
Researchers further utilized whole-genome sequencing to identify the molecular mechanisms contributing to Huanglongbing (HLB) tolerance in a sweet orange mutant. The results showed that although the infections of disease-tolerant T19 were equivalent to those of sensitive T78 and DVS, T19 trees had a higher leaf area index and healthier growth.
The T19 trees and a lost-tag specimen SF confirmed to be genetically identical to T19 displayed no somatic structural variations (SVs) and shared three somatic SVs. These variations involved complex translocations and insertions, affecting several genes and potentially contributing to HLB tolerance. Further investigation into allelic expression showed significant upregulation and downregulation of alleles in T19 compared to DVS and T78.
There was a significant change in the expression ratio of biased alleles and alleles in DVS, T19, and T78. T19 demonstrated unique upregulation patterns in stress response genes, mitochondrial metabolism, and other areas not observed in DVS or T78. This was supported by transcriptomic profiling and the presence of differentially expressed microRNAs in T19.
Moreover, T19 had significantly more upregulated heat shock proteins (HSPs), which are crucial for stress tolerance, and genes involved in reducing reactive oxidative species, which are known to alleviate HLB symptoms. The findings suggested that these transcriptomic differences, likely resulting from somatic mutations, underlie the superior HLB tolerance in T19, primarily through enhanced stress responses and protection of phloem protein homeostasis.
In conclusion, researchers have assembled a high-precision phased diploid genome of Valencia sweet orange, enabling unprecedented allele-level studies in citrus genetics. This genomic assembly not only clarifies the complex genetics of sweet oranges but also promises to guide future genetic engineering and breeding programs for improved citrus varieties.
More information: Bo Wu et al, A chromosome-level phased genome enabling allele-level studies in sweet orange: a case study on citrus Huanglongbing tolerance, Horticulture Research (2022). DOI: 10.1093/hr/uhac247
Journal information: Horticulture Research
Provided by NanJing Agricultural University





















