This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Decoding the Cascade hop genome: Unraveling evolutionary secrets and divergence in the Cannabaceae family
Hop (Humulus lupulus L. var. Lupulus) is a diploid, wind-pollinated, perennial plant. Not only does the H. lupulus play an important role in brewing and flavoring, but its female inflorescences contain lupulin glands (glandular trichomes) that can synthesize and store resins, bitter acids, essential oils, and flavonoids.
Hop cultivar Cascade is widely produced in America, known for its floral and citrus aroma. The optimal latitude for hop growth falls between 35° and 55° in the Northern and Southern hemispheres. Humulus and Cannabis both belong to the Cannabaceae family, but ongoing debate exists about their divergence dates.
With the development of new genomic data, it is possible to re-evaluate and refine divergence dates. The hop genome is large and heterozygous, and even with long-read sequencing, the assembly remained fragmented (N50 ∼ 673 kb) and contig order was unknown. Therefore, contiguous genome sequences are necessary to study synteny and genomic organization.
Horticulture Research published a perspective entitled "An improved assembly of the "Cascade" hop (Humulus lupulus) genome uncovers signatures of molecular evolution and refines time of divergence estimates for the Cannabaceae family".
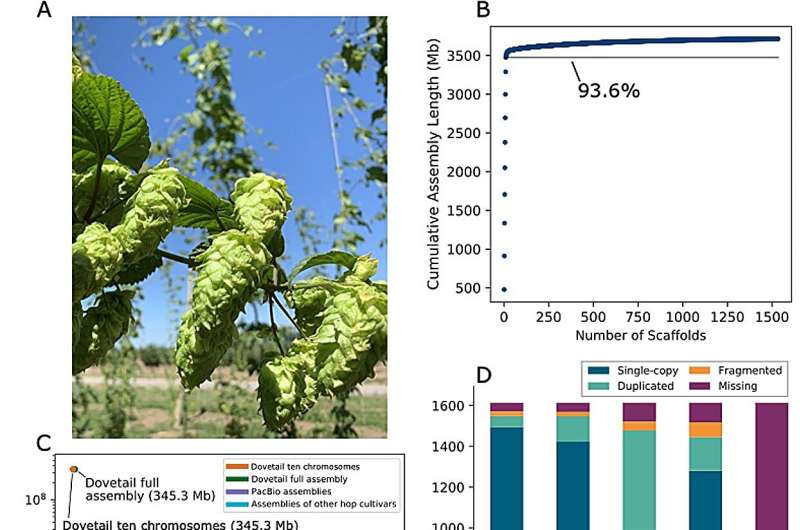
In this study, the researchers describe an improved Cascade female genome assembly and the corresponding analysis of genome content, organization, and evolution. Initially, the size of the PacBio primary assembly used to anchor the scaffolds was approximately 3.71 Gb, which was then further improved using the Dovetail Hi-C assembly. Notably, the N50 increased from 673 kb (using PacBio) to 345.208 Mb (using Dovetail), reflecting a more contiguous assembly.
Some 93.6% of the assembly was contained in the largest 10 scaffolds, which likely correspond to the 10 chromosomes of the hop genome. GC content of the full, polished Dovetail assembly was 39.13% and 0.022% Ns. The results showed an enrichment of CHH trinucleotides associated with DNA methylation, which is involved in gene regulation of essential plant processes.
Evaluating assembly completeness using BUSCO, it was noted that assembly statistics improved post-polishing, showing greater alignment with previous hop genome research. The genome size was estimated to be approximately 3.058 Gb, with ∼4.59%–5.47% heterozygous and 64.25% repeat sequences. The genetic map produced 10 linkage groups. Average genetic distance between markers is 0.35 cM with an average of 409 markers per linkage group.
Then, researchers utilized a combination of Transdecoder and MAKER to generate gene models. Among the gene models produced by Transdecoder, 94.9% were found on the top 10 scaffolds. Gene model completeness was assessed using BUSCO.
Repeat-associated gene models were identified and removed based on similarity to Pfam domains and UniProt genes. A total of 23,583 genes showed similarity to a UniProt Embryophyta gene. After excluding repeat-associated gene models and MAKER gene models without known similarities, 30,404 gene models remained for further synteny, orthology, and evolution studies. By comparing protein sequences of hop to seven other plant species, 22,739 orthologous gene groups (OGGs) were identified with OrthoFinder.
Furthermore, the densities of genes and long terminal retrotransposon (LTR) on the 10 largest scaffolds were visualized. The circos plot showed that in most scaffolds, gene density was higher at the end. Notably, the third largest scaffold contained most significant sex-associated markers, suggesting its role as a putative X chromosome. Syntenic gene blocks were depicted in the center track of the circos plot, with most syntenic blocks occurring within the same scaffold.
Comparisons of hop and hemp genomes revealed extensive sequence similarities. Large genomic sequences unique to hop chromosomes were also observed. In addition, defense and terpene genes' occurrences in syntenic blocks were analyzed, indicating chromosomes 8 would be a useful target scaffold for further investigation of defense genes. Molecular evolution analysis using Ks distributions revealed that hop and hemp diverged at 16.013 mya.
A fossil-calibrated time tree was computed, identifying a divergence time for Humulus and Cannabis at approximately 22.6438 million years ago. Finally, an orthologous gene analysis revealed expanded and contracted hop gene families, shedding light on functional enrichments in these groups.
In conclusion, this groundbreaking study sets the stage for deeper insights into hop evolution, defense response and metabolism. The Cascade chromosome-level assembly will also guide future studies in gene organization, structural variations among hop cultivars, and broader Cannabaceae genomics.
More information: Lillian K Padgitt-Cobb et al, An improved assembly of the "Cascade" hop (Humulus lupulus) genome uncovers signatures of molecular evolution and refines time of divergence estimates for the Cannabaceae family, Horticulture Research (2022). DOI: 10.1093/hr/uhac281
Journal information: Horticulture Research
Provided by NanJing Agricultural University



















