This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Chromosome-scale genome sequence of Suaeda glauca sheds light on salt stress tolerance in halophytes
A research paper titled "Chromosome-scale genome sequence of Suaeda glauca sheds light on salt stress tolerance in halophytes," by Professor Qin Yuan's team from the Center for Genomics, Haixia Institute of Science and Technology (Future Technology College) at Fujian Agriculture and Forestry University, has been published in "Horticulture Research".
Soil salinity is a growing concern for global crop production and the sustainable development of humanity. Therefore, it is crucial to comprehend salt tolerance mechanisms and identify salt-tolerant genes to enhance crop tolerance to salt stress.
S. glauca, a halophyte species well-adapted to the seawater environment, possesses a unique ability to absorb and retain high salt concentrations within its cells, particularly in its leaves, suggesting the presence of a distinct mechanism for salt tolerance.
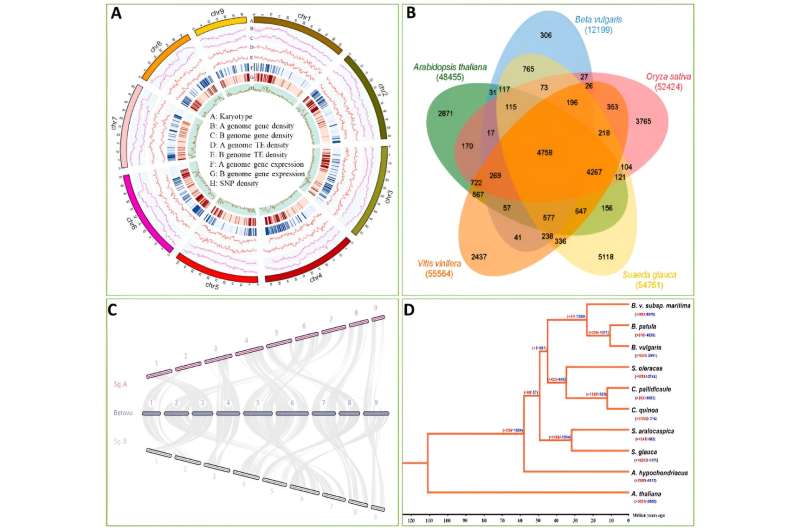
In this study, the authors performed de novo sequencing of the S. glauca genome. The genome has a size of 1.02 Gb (consisting of two sets of haplotypes) and contains 54,761 annotated genes, including alleles and repeats.
Comparative genomic analysis revealed a strong synteny between the genomes of S. glauca and B. vulgaris. Of the S. glauca genome, 70.56% comprises repeat sequences, with Retroelements being the most abundant.
Leveraging the allele-aware assembly of the S. glauca genome, we investigated genome-wide allele-specific expression in the analyzed samples. The results indicated that the diversity in promoter sequences might contribute to consistent allele-specific expression (refer to full text for details).
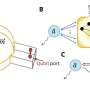
The typical angiosperm flower consists of four whorls: sepal, petal, stamen, and carpel, although deviations from this arrangement can occur in some species. In S. glauca, only three whorls were observed, with the first whorls significantly degraded and the second whorls exhibiting sepal-like characteristics. The third and fourth whorls show the normal phenotype.
To confirm the sepal-like identity of the second whorl of the S. glauca flower, the researchers analyzed the expression of genes involved in photosynthesis and chlorophyll synthesis-related pathways.
The clustered heat maps showed that the expression patterns of the genes examined in the second whorls were similar to those of the sepals. Moreover, a systematic analysis of the ABCE gene family shed light on the formation of S. glauca's flower morphology, suggesting that the dysfunction of A-class genes is responsible for the absence of petals in S. glauca.
In order to gain an in-depth understanding of the salt tolerance mechanisms in Suaeda genus plants from a genomic perspective, they conducted a comparative genome analysis between Suaeda species, including Suaeda aralocaspica (a salt-tolerant plant within the Amaranthaceae family) and other non-salt-tolerant species from the same family.
-
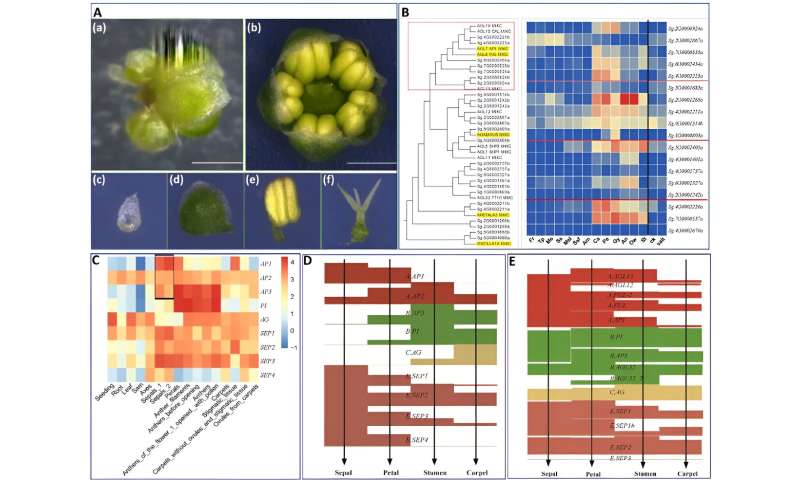
Flower morphology and ABCE genes of S. glauca. Credit: Horticulture Research -
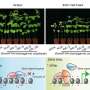
Strategy and results of ortholog expansion analysis of S. glauca. Credit: Horticulture Research -
Time-course RNA-seq analysis of S. glauca roots and leaves under salt treatment. Credit: Horticulture Research
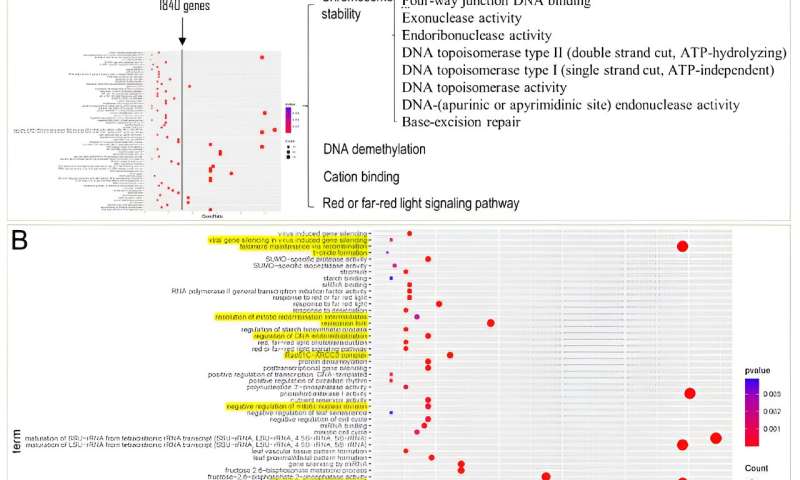
The results revealed a significant expansion of gene families in these two salt-tolerant Suaeda species, encompassing a total of 1,840 genes. Enrichment analysis of these genes unveiled a notable enrichment of Gene Ontology (GO) terms related to DNA repair, chromosome stability, DNA demethylation, cation binding, and red/far-red light signaling pathways within the shared expanded gene families of Suaeda species.
In comparison to non-halophytic species in the Amaranthaceae family, the expanded genes in Suaeda species are predominantly enriched in the "DNA/chromatin stability maintenance" function. This suggests that the role of "DNA/chromatin stability maintenance" may play a crucial role in the salt tolerance of Suaeda plants.
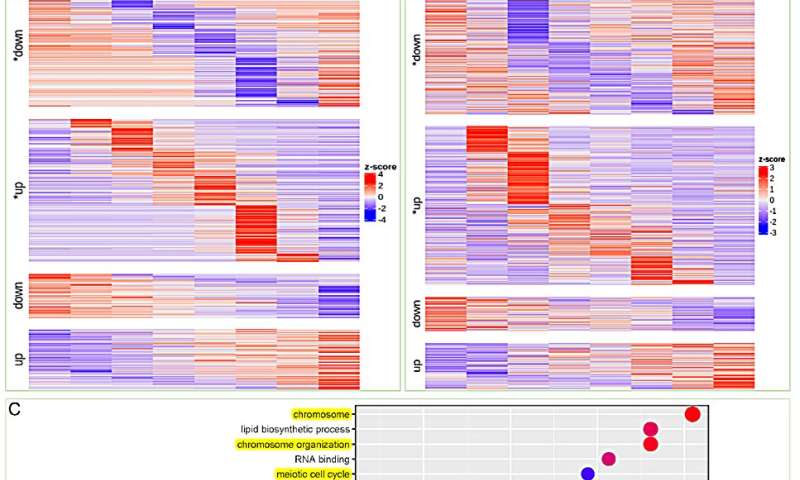
To further validate the aforementioned observations, the researchers conducted a temporal RNA-seq analysis on salt-treated Suaeda plants. Cluster analysis of differentially expressed genes at various time points following salt treatment was shown. Notably, the transition-up genes were identified as a stable response mechanism for S. glauca to cope with salt stress.
Interestingly, the transition upregulated genes in leaves were mainly associated with DNA repair and chromosome stability, along with GO enrichment related to lipid biosynthetic process and isoprenoid metabolic process. These results are consistent with the previous analysis of the S. glauca gene family expansion.
Additionally, genome-wide analysis of transcription factors indicated a significant expansion of the FAR1 gene family (refer to full text for details). However, further investigation is needed to determine the exact role of the FAR1 gene family in salt tolerance in S. glauca.
In this study, we de novo assembled the haploid genome of S. glauca at the chromosomal level, say the researchers. The genome sequence is of high quality, and the annotation is extensive, establishing it as a reference genome for the halophyte S. glauca.
Additionally, we elucidated a novel salt tolerance mechanism involving "DNA/chromatin stability maintenance" in halophytic plants. To our knowledge, this mechanism of salt tolerance is a recent discovery within halophytic plants, potentially offering new insights and avenues for enhancing crop salt tolerance, they said.
More information: Yan Cheng et al, Chromosome-scale genome sequence of Suaeda glauca sheds light on salt stress tolerance in halophytes, Horticulture Research (2023). DOI: 10.1093/hr/uhad161
Provided by NanJing Agricultural University