This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Decoding the 'montmorency' sour cherry genome: Unraveling prunus evolution and enhancing breeding strategies
The sour cherry (Prunus cerasus L.) is a treasured temperate fruit, known for its distinct sweet-acidic flavor and exceptional processing quality. Belonging to the economically vital Rosaceae family, it shares lineage with various Prunus species such as peach, sweet cherry, and almond.
Tracing the evolutionary journey of Prunus has historically been challenging, given the widespread hybridization and polyploidy. Studies have identified sour cherry as an allotetraploid (2n = 4x = 32), a result of a hybridization between Prunus fruticosa Pall. and Prunus avium L. Furthermore, climatic challenges like early spring freezes pose significant threats to the temperate fruit industry, causing massive crop loss.
Addressing this, Michigan State University's sour cherry breeding program aims to comprehend the genetic control of bloom time. However, the absence of a public sour cherry genome sequence has limited these studies to traditional linkage maps and marker methods. Therefore, developing a sour cherry genome resource is essential for supporting breeding efforts, enabling gene discovery, and understanding the genetic foundation of agronomic traits in this complex tetraploid.
In May 2023, Horticulture Research published a research paper entitled "Genome of tetraploid sour cherry (Prunus cerasus L.) 'Montmorency' identifies three distinct ancestral Prunus genomes."
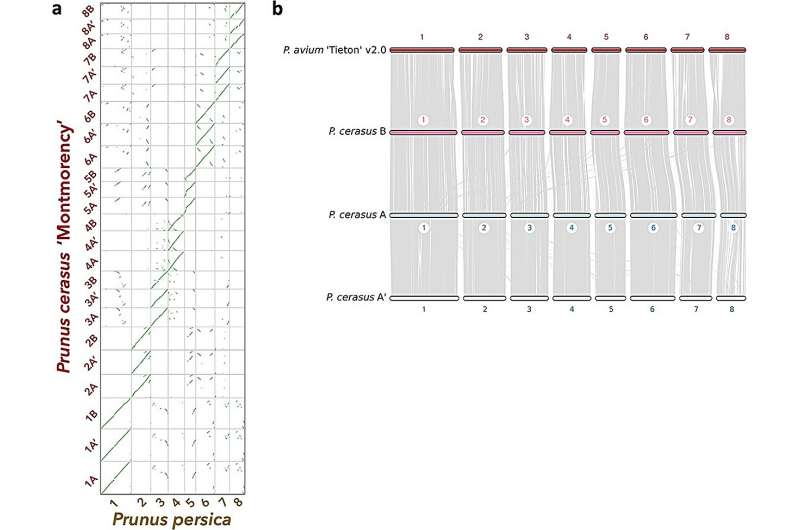
Researchers first assembled the sour cherry "Montmorency" genome using a combination of PacBio and Illumina sequencing, further scaffolded with chromosome conformation capture (Hi-C). Although expecting two full haplotypes due to the cherry's allotetraploid nature, they discovered 24 linkage groups. Eight of these hinted at origins from the P. avium-like ancestor.
Further analyses, including k-mer clustering, segregated these chromosomes into three distinct sets named A, A', and B, representing different subgenomes derived from separate Prunus species. An examination of the Illumina reads revealed that subgenome B had twice the genome dosage compared to A or A', confirming the "Montmorency" genome structure as AA'BB.
For structural annotation of gene space, the researchers predicted 92,783 protein-coding genes using RNA sequencing and long-read cDNA datasets from the "Montmorency" tissue. With further analysis using defusion, they found 906 potentially fused genes in the full assembly, of which 707 were present in the scaffolded assembly.
Then, 4,481 genomic regions on scaffold components were locally relabeled using MAKER within defusion, and 9,777 defused gene models obtained through another Pfam domain search were also re-added to the final annotation. The annotated genes had high BUSCO completion scores and displayed an AED distribution suggesting a high-quality annotation.
For Prunus fruticosa, a probable allotetraploid, a draft genome was assembled using PacBio and Illumina reads. Analysis indicated that P. fruticosa is also an allotetraploid, with its assembly showcasing common issues linked to polyploid assemblies. Its annotation predicted 102,361 protein-coding genes. Progenitor assignment indicated "Montmorency" subgenomes A and A' as P. fruticosa-like, while subgenome B was P. avium-like.
In addition, the Dormancy-Associated MADS-box genes (DAMs), essential for Prunus dormancy and blooming, were analyzed in "Montmorency" and P. fruticosa. Using BLAST+ and manual adjustments via Apollo v 2.6.5, researchers identified three full DAM1–DAM6 haplotypes in "Montmorency."
One, DAM6 on chr1A', had a unique intron-exon structure. P. fruticosa exhibited two contigs with full DAM haplotypes, mirroring those in "Montmorency." Additionally, four S-alleles, associated with Prunus self-incompatibility, were detected in "Montmorency" with over 99% sequence resemblance to prior studies.
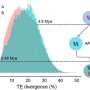
P. fruticosa revealed an S-allele akin to P. cerasus S36b, suggesting "Montmorency" sour cherries derived their S36 from a P. fruticosa ancestor. The research also indicated that P. cerasus "Montmorency" evolved from a hybridization event less than 1.61 million years ago.
In conclusion, this study presents a detailed chromosome-scale genome assembly for the predominant U.S. sour cherry cultivar, "Montmorency," revealing its trigenomic composition of AA'BB. Additionally, the genome of P. fruticosa, a key ancestor, was assembled and utilized alongside a P. avium sequence.
Key gene sets, including the Dormancy-Associated MADS-box genes (DAMs) and self-incompatibility S-alleles, were annotated. This research enriches our understanding of Prunus evolution and offers crucial insights for breeding and comparative genomics, promising advancements in sour cherry cultivation and addressing issues like low fruit set.
More information: Charity Z Goeckeritz et al, Genome of tetraploid sour cherry (Prunus cerasus L.) 'Montmorency' identifies three distinct ancestral Prunus genomes, Horticulture Research (2023). DOI: 10.1093/hr/uhad097
Journal information: Horticulture Research
Provided by NanJing Agricultural University