This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Researchers provide first chromosome-level genome assembly in Murraya plants
Murraya paniculata, commonly known as orange jessamine in the family Rutaceae, is an important ornamental plant in tropical and subtropical regions that is famous for its strong fragrance. As an ornamental plant with strong flower aroma, the floral volatiles of M. paniculata differ from those of Citrus plants in the Aurantioideae. However, the molecular basis for the biosynthesis of the compounds and the reason for the significant differences in volatiles between Citrus and Murraya are still unknown.
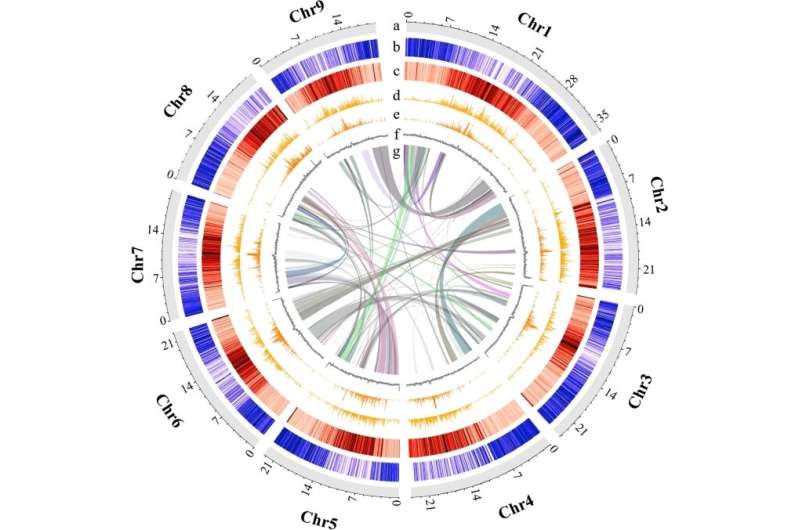
In a study published in BMC Biology, researchers from the Xishuangbanna Tropical Botanical Garden (XTBG) and the Kunming Institute of Botany of the Chinese Academy of Sciences reported the first chromosome-level reference genome of M. paniculata with a total assembly length of 216.87 Mb from 9 chromosomes and a contig N50 of 18.25 Mb.
The researchers performed comparative analyses with other published Aurantioideae genomes and found that transposon expansion was responsible for the difference in genome size between M. paniculata and the other Aurantioideae species.
Obvious differences in transposon content were detected in the genomes of M. paniculata and other Aurantioideae species, especially in the upstream regions of genes that may affect metabolite biosynthesis.
They then carried out the analysis of flower volatiles and transcriptome analysis in M. paniculata and C. maxima at different developmental stages. And they identified three new genes with strong synthetic activity for phenylacetaldehyde in M. paniculata. The expressions of two homologues of these genes in C. maxima may be affected by long terminal repeat (LTR) insertions, which resulted in the absence of phenylacetaldehyde in C. maxima flowers.
"We have presented a high-quality reference genome of M. paniculata, which is the first chromosome-level genome assembly in Murraya plants. Our study provides insights into how transposons contribute to the variation in flower volatiles between Murraya and Citrus plants," said Prof. Yang Yongping of XTBG.
More information: Tianyu Yang et al, Chromosome-level genome assembly of Murraya paniculata sheds light on biosynthesis of floral volatiles, BMC Biology (2023). DOI: 10.1186/s12915-023-01639-6
Journal information: BMC Biology
Provided by Chinese Academy of Sciences




















