This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Researchers publish the first grape T2T reference genome
A new article titled "The complete reference genome for grapevine (Vitis vinifera L.) genetics and breeding" has been published in Horticulture Research.
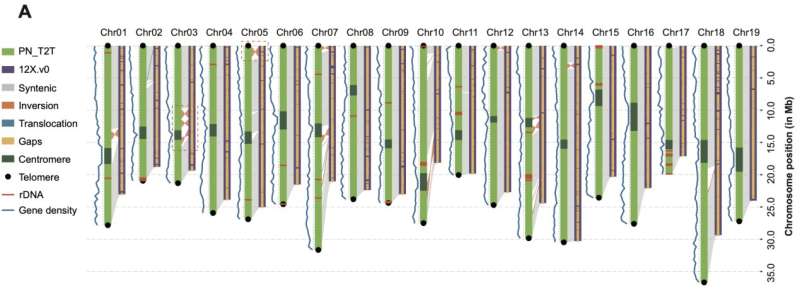
This gap-free PN_T2T genome (494.87 Mb) has improved significantly over previous versions. The contiguous N50 length of PN_T2T was ~ 250 times higher than that of 12X.v0 (25.93 Mb versus 102 kb), and all the 9429 gaps in 12X.v0 were filled in PN_T2T genome. Orientation errors in 12X.v0 were also corrected such as inversions and translocations compared to PN_T2T.
The authors found the telomere repeat unit (TTTAGGG/CCCTAAA) was the most abundant in both ends of each chromosome. As for centromeric regions, the authors found 107 bp repeats were the most abundant unit in the whole genome, which had 182,620.5 (copies ≥ 2) repetitions accounted for about 3.95% of the genome. A total of 343 genes were captured in the centromeres, and RNA modification, protein autophosphorylation, DNA integration, DNA recombination and photomorphogenesis appeared enriched while exploring biological process (BP)-related terms of these genes.
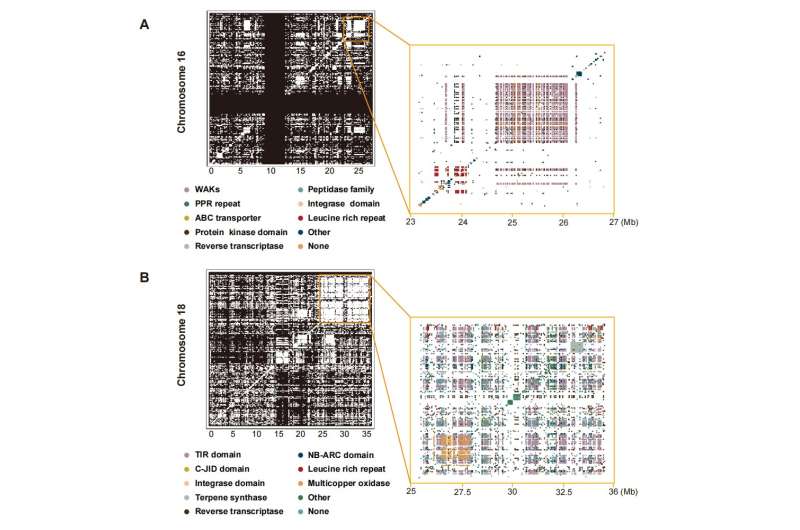
The authors found a total of 377 gene clusters in the grapevine reference genome; these duplications often involve local rearrangements and can extend into megabases with dozens to hundreds of genes involved. On chromosome 16 (23-27 Mb), there were 599 enriched domain genes; on chromosome 18 (25~36 Mb), there were 1,237 genes enriched domains. Among them, many strongly enriched domains are part of plant disease resistance base (R base) domains, and these R genes and gene clusters in grapes highlight a tremendous opportunity for exploring plant defense mechanisms.
Although the PN40024 genome is highly homozygous (99.8%), combined with the resequencing data of PN40024, the authors found nine hotspots of heterozygous SNPs on chromosomes. The results showed that the most significantly enriched terms were response to water deprivation, protein phosphorylation, cell division, response to oxidative stress and response to salt stress, which were closely associated with key physiological activities in plants.
Overall, the previous versions of the grapevine reference genome consisted of thousands of fragments with missing centromeres and telomeres, which limited the accessibility of the heredity of important agronomic traits in these regions. However, this gap-free reference genome, the first T2T reference genome for fruit trees for the PN40024 genome, provides important resources for grapevine genetics and breeding.
More information: Xiaoya Shi et al, The complete reference genome for grapevine (Vitis vinifera L.) genetics and breeding, Horticulture Research (2023). DOI: 10.1093/hr/uhad061
Provided by NanJing Agricultural University