This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Complete chloroplast genomes clarify phylogenetic placement of six polygonatum species
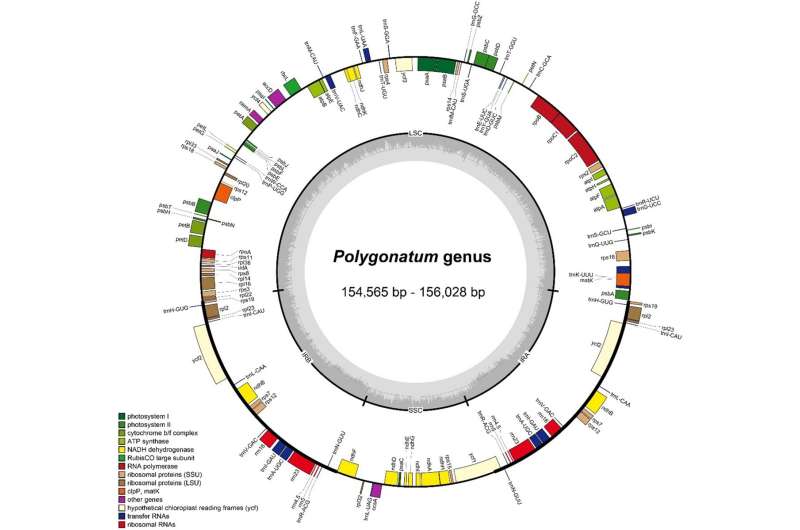
Polygonatum Miller is highly valued for its medicinal properties with the horizontal creeping fleshy roots. Previous studies have mainly focused on the size and gene content of the plastid genome, with insufficient studies on the comparative genomic analysis. In addition, the complete chloroplast genome data of some species have not been published, so their phylogenetic placement is not well understood.
A research team led by Prof. Hu Guangwan from the Wuhan Botanical Garden of the Chinese Academy of Sciences reported the first complete chloroplast genome of P. campanulatum, and then conducted comparative and phylogenetic analysis of the complete chloroplast genomes of six Polygonatum species.
The chloroplast genome showed a typical quadripartite structure, with the length between 154,564 and 156,028 bp in Polygonatum. A total of 113 unique genes were detected in each of the species. Comparative analysis revealed that the genome size, gene content, gene order and guanine-cytosine content maintained a high similarity in the chloroplast genomes of Polygonatum and Heteropolygonatum.
All species showed no significant contraction or expansion of IR boundaries, except for P. sibiricum1, in which the rps19 gene turned out to be a pseudogene, due to the incomplete duplication. Abundant long dispersed repeats and simple sequence repeats were detected in each genome.
Five highly variable regions were found to be potential specific DNA barcodes, and 14 genes were revealed to be under positive selection and a large number of repetitive sequences were identified.
62 chloroplast sequences of Polygonatum and its related species were used for phylogenetic analyses. The results showed that Polygonatum could be divided into two significant clades, sect.Verticillata and sect. Sibirica plus sect. Polygonatum.
Furthermore, P. campanulatum and P. tessellatum + P. oppositifolium were strongly supported to be sister species and placed in sect. Verticillata, suggesting that leaf arrangement does not appear to be suitable basis for delimiting of subgeneric groups in Polygonatum.
Additionally, P. franchetii was also sister to P. stenophyllum within sect. Verticillata as well.
This study provides more chloroplast genomic information of Polygonatum and contributes to improve species identification and phylogenetic studies in further work.
The paper is published in the journal Scientific Reports.
More information: Dongjuan Zhang et al, Comparative and phylogenetic analysis of the complete chloroplast genomes of six Polygonatum species (Asparagaceae), Scientific Reports (2023). DOI: 10.1038/s41598-023-34083-1
Journal information: Scientific Reports
Provided by Chinese Academy of Sciences



















