This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Decoding cherry traits: Genome-wide association studies propel sweet cherry breeding
Sweet cherry (Prunus avium L.) breeding has advanced through the identification of molecular markers for key traits, with genome sequencing of leading cultivars providing a foundation for marker-assisted selection (MAS).
Despite this progress, the resolution of quantitative trait locus (QTL) analyses has been inadequate, often missing gene associations. The traditional breeding process is also hampered by low efficiency and high costs.
Recent strides in whole-genome sequencing have revealed nearly 2 million single nucleotide polymorphisms (SNPs), offering higher marker density for genome-wide association studies (GWAS). However, the distribution of SNPs remains uneven, hindering the pinpointing of candidate genes. The current challenge lies in harnessing the potential of GWAS for high-resolution mapping to expedite the breeding of new, improved sweet cherry varieties.
In October 2022, Horticulture Research published a perspective titled "High-resolution genome-wide association study of a large Czech collection of sweet cherry (L.) on fruit maturity and quality traits".
In this study, researchers first utilized paired-end sequencing on the Illumina NovaSeq 6000 platform, which produced a 4.15-Tb sequence with individual samples contributing 7.6–36.8 Gb. This sequence depth provided extensive genome coverage, allowing for the identification of 1,767,106 high-quality single nucleotide polymorphisms (SNPs) distributed across the accessions.
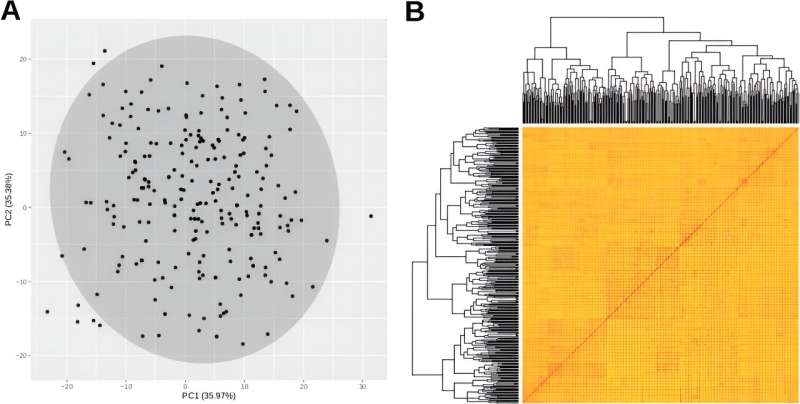
In-depth analysis using principal component analysis (PCA) and Bayesian information criterion (BIC) revealed no subpopulations or strong sample clustering within the genetic structure, indicating a homogenous population without distinct genetic subgroups.
Phenotypic diversity was assessed from 2012 to 2020, unveiling significant variability across various traits, more pronounced within the Collection of Genetic Resources (CGR) accessions than in the breeding materials (BM). Color traits notably differed between the populations, with the CGR accessions displaying a broader color spectrum. Size traits such as fruit weight were larger in the BM accessions, correlating with the goals of selective breeding which also tend towards later ripening characteristics.
In addition, the genome-wide association study (GWAS) using the FarmCPU model for its close-to-ideal genomic inflation factor revealed 59 SNPs associated with eleven traits, including time of harvest, where specific SNP genotypes corresponded with early or late ripening. Similarly, 10 SNPs were found to be significantly associated with fruit and flesh color, chr3_23939472 and chr1_ 41003304 were related to both fruit color and flesh color.
A critical discovery was the identification of a deletion in the MYB10 region, which is involved in fruit color determination, with yellow fruits having a homozygous deletion in this region. Moreover, the study found a significant association between 18 SNP markers and the firmness traits of the fruits, indicating genetic underpinnings for the textural qualities of cherries. Likewise, 26 SNPs were associated with fruit size and weight, pinpointing the genetic basis for these commercially important traits.
Finally, haplotype blocks were identified that encompass significant SNPs co-inherited within the population, aiding in the mapping of candidate genes associated with specific traits. Out of 141 candidate genes identified, 104 had known functions, offering insights into their potential roles in determining cherry phenotypes.
Although no subgroups were identified in the principal component analysis of 299 samples (CGR-BM), slight differences were found between BM and CGR. By comparing the positions of the genomes, it was confirmed that 22 SNPs co-located with known QTLs were associated with loci previously described as QTLs for specific or similar traits.
In conclusion, a total of 1,767,106 single nucleotide polymorphisms (SNPs) were discovered, offering a detailed genetic map. A highly significant set of SNPs was identified in the sweet cherry material population, which were related to harvest time, fruit color, hardness and size, and haplotype blocks.
These findings enhance the understanding of the genetic determinants of key phenotypical traits in sweet cherry and provide a solid foundation for future breeding efforts.
More information: Kateřina Holušová et al, High-resolution genome-wide association study of a large Czech collection of sweet cherry (Prunus avium L.) on fruit maturity and quality traits, Horticulture Research (2022). DOI: 10.1093/hr/uhac233
Journal information: Horticulture Research
Provided by NanJing Agricultural University