This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
proofread
New single-nucleotide polymorphism chip that can help identify genetic markers of desirable traits in rice
Genetic markers such as fragment length polymorphisms (RFLP), simple sequence repeats (SSRs), and single-nucleotide polymorphisms (SNPs) provide unique identifiers for individual organisms. This aids the identification of significant genetic variations in plants, allowing modern plant breeding to select superior crop varieties. Next-generation sequencing (NGS) has enhanced marker-assisted selection or backcross breeding of crops, which is the transfer of a desired trait into the favored genetic background of another genome.
However, due to its expensive nature and extensive data processing requirements, NGS is not practical for screening large populations of crop plants, especially among small and medium breeders. A more cost-effective and efficient solution to assess desirable genetic traits in these populations is SNP arrays. These chips have embedded DNA probes that interact with a genetic sample, enabling the detection of hundreds of thousands of variations among two or more plant samples.
Tapping the potential of this approach, Associate Professor Wensheng Wang from the Chinese Academy of Agricultural Sciences and his research team have created a SNP array containing more than 56,606 genetic markers for rice.
"Globally, more than 2.6 billion people rely on rice as a staple in their diet. The gene chip developed in our study can be an effective tool for breeders in selecting superior, new, high-quality rice varieties that are more flavorful, better tasting, salt tolerant, and resistant to diseases and insects," shared Wang. "Moreover, the gene chip can be used for various genetic studies."
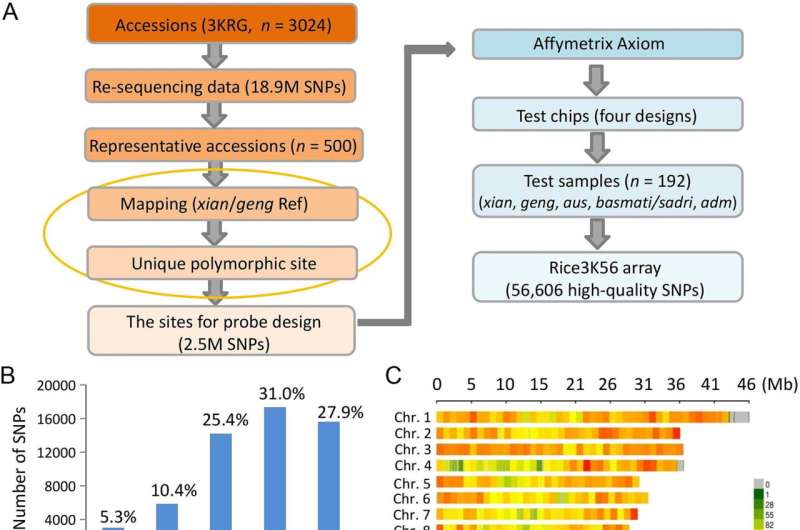
The researchers developed the custom SNP array by collecting and sequencing genetic samples belonging to 3,024 rice samples from around the world. From more than 18.9 million high-quality SNPs from this sample, approximately 2.5 million polymorphic SNPs were identified and retained in the array design. These SNPs were then printed on a genotyping chip platform (Affymetrix) with four designs.
The effectiveness of the array was tested by using a representative set of 192 rice varieties, and the results were impressive. On average, it correctly identified 99.6% of the relevant genome, while also demonstrating high density and uniform coverage with an average distance of 6.7-kb between two adjacent SNPs. Furthermore, the data obtained from this platform offered more genetic variability information than other previously developed arrays. The team reported their findings in The Crop Journal.
"Rice3K56 can help small and medium-sized companies select for superior rice varieties in a manner that is less subjective and low-effort," said Wang. "Our work makes genotyping more mechanized and streamlined, resulting in higher efficiency and accuracy. In the long run, this can ensure not only food security but also a diversification of rice flavors."
More information: Chaopu Zhang et al, Rice3K56 is a high-quality SNP array for genome-based genetic studies and breeding in rice (Oryza sativa L.), The Crop Journal (2023). DOI: 10.1016/j.cj.2023.02.006
Provided by KeAi Communications Co., Ltd.