This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Unlocking the future of apple breeding: The power of genomewide selection in modern breeding practices
Apple (Malus domestica Borkh.) breeding is a resource-intensive and time-consuming process. Although a large number of quantitative trait loci (QTLs) associated with apple fruit appearance and quality traits have been reported, fewer QTLs have been translated into locus-specific traits for use in breeding, while many apple fruit quality traits are genetically complex and may be influenced by many minor-effect QTLs throughout the genome.
In addition, the long juvenile period of apples, coupled with the challenges of phenotyping, exacerbates the complexity of the problem. Therefore there is an urgent need to find ways to improve breeding efficiency, but research on its application in apple breeding through genomewide selection (genomic selection) is still scarce.
In May 2023, Horticulture Research published a research paper entitled "Genomewide selection for fruit quality traits in apple: breeding insights gained from prediction and postdiction."
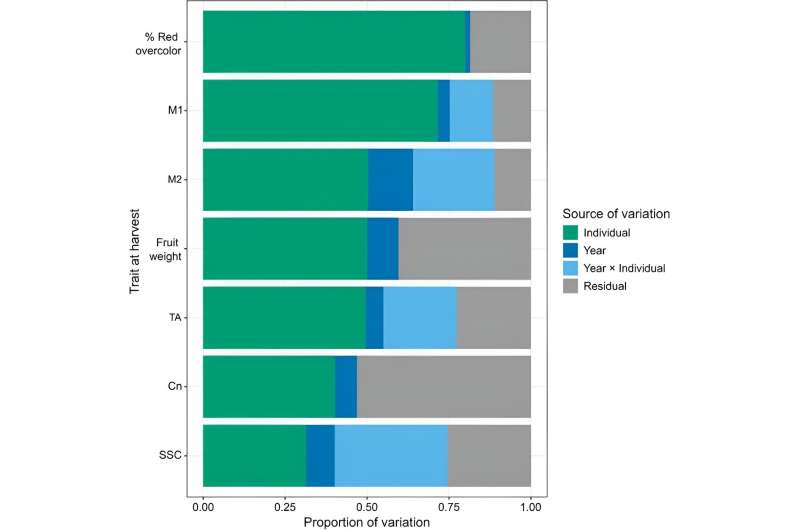
In this study, fruit quality traits were investigated to understand trait variation and predictive ability. Firstly, both year (p
Quantitative differences were observed between individuals in the germplasm set. Quantitative variation was observed among individuals in the germplasm set. Next, the study predicted quality traits for fruit at harvest.
The results showed that the larger the training set, the higher the prediction ability, with M1 having the highest prediction ability (mean = 0.56) and Cn the lowest (mean = 0.36). The effect of single nucleotide polymorphisms (SNPs) per chromosome of individual family on predictive ability was then investigated. The number of SNPs per chromosome, trait, and test family had a significant effect on predictive abilities.
The largest increase in predictive ability was observed when comparing 10 SNPs to five SNPs per chromosome. When predicting individual offspring within families, predictive abilities varied significantly by family and trait. Across traits, larger families (with more phenotyped offspring) generally showed higher predictive ability than smaller ones. Also, the inclusion of SNPs at specific quantitative trait loci (QTLs) as fixed effects sometimes improved predictive abilities, especially noticeable for traits like red over-color.
Lastly, genomewide postdiction was considered. Here, fewer offspring were culled based on predicted values (genomewide selection) than based on observed trait values (phenotypic selection). When different culling thresholds were applied, the number of culled individuals varied, with thresholds based on "Honeycrisp" trait values culling the fewest individuals. Conversely, thresholds derived from percentile values often culled the most, leading to more disagreement between phenotypic and genomewide selection decisions.
In conclusion, this study explored the effectiveness of genomewide selection in predicting fruit quality traits in apple breeding, especially in the context of the UMN apple breeding program. This research underscores the potential of genomewide selection as a powerful tool for apple breeding and provides a promising avenue for future applications in enhancing fruit quality.
More information: Sarah A Kostick et al, Genomewide selection for fruit quality traits in apple: breeding insights gained from prediction and postdiction, Horticulture Research (2023). DOI: 10.1093/hr/uhad088
Journal information: Horticulture Research
Provided by NanJing Agricultural University