Grapevine breeding programs assisted by genomic prediction

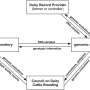
Breeding schemes typically involve first the choice of parents and then the selection of offspring within crosses. Genomic prediction is adapted both for predicting the cross mean and for ranking the genotypes within a cross. These steps correspond to the components of the predictive ability of genomic prediction. The cross mean is the sum of the breeding values of the parents if allelic effects are only additive, but in practice, some deviation may result from dominance or epistasis. To date, only a few studies have investigated cross mean predictive ability in heterozygous crops, and none have investigated the parameters that influence it.
Very few authors have assessed the potential utility of genomic prediction in grapevine (Vitis vinifera subsp. vinifera). Before genomic selection can be used in grapevine, predictive ability must be evaluated across populations. In particular, predictive ability can be evaluated using a diversity panel and a bi-parental progeny as training and validation sets, respectively. This is a challenging configuration, given the low genetic relatedness, but this configuration is much more likely to occur in actual breeding schemes than genomic prediction within the same population. As in grape, studies investigating across-population genomic prediction are also lacking for most clonally propagated crops.
Recently, scientists from IFV-INRAE-Institut Agro tested across-population genomic prediction in a more realistic breeding configuration. They evaluated prediction quality over 15 traits of interest (related to yield, berry composition, phenology, and vigor) for both the average genetic value of each cross (cross mean) and the genetic values of individuals within each cross (individual values). Genomic prediction under these conditions was found to be useful: for cross mean, the average per-trait predictive ability was 0.6. Per-cross predictive ability was halved on average, but reached a maximum of 0.7. The mean predictive ability for individual values within crosses was 0.26, about half the within-half-diallel value taken as a reference. For some traits and crosses, these across-population predictive ability values are promising for implementing genomic selection in grapevine breeding.
"We implemented genomic prediction in grapevine in a breeding context, i.e. across populations, on 15 traits, in ten related crosses, and obtained moderate to high PA values for some crosses and traits, thus showing genomic prediction usefulness in grapevine. Never before had genomic prediction been implemented for so many traits and crosses simultaneously in this species," Dr. Agnès Doligez said. These results will greatly help to design grapevine breeding programs assisted by genomic prediction.
The research was published in Horticulture Research.
More information: Charlotte Brault et al, Across-population genomic prediction in grapevine opens up promising prospects for breeding, Horticulture Research (2022). DOI: 10.1093/hr/uhac041
Provided by Nanjing Agricultural University The Academy of Science