This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Scientists uncover surprising twist in the ways bacteria spread antibiotic-resistant genes
Scientists have found a counterintuitive wrinkle in the way bacteria spread antibiotic-resistant genes through small circular pieces of DNA called plasmids.
Plasmids, found in bacteria and some other microorganisms, are physically separate from chromosomal DNA and can replicate on their own. Bacteria can acquire plasmids from other bacterial cells or from viruses, and as plasmids build up, they give bacteria antibiotic resistance.
But some plasmids are easier for bacteria to acquire than others. What makes these plasmids spread more easily?
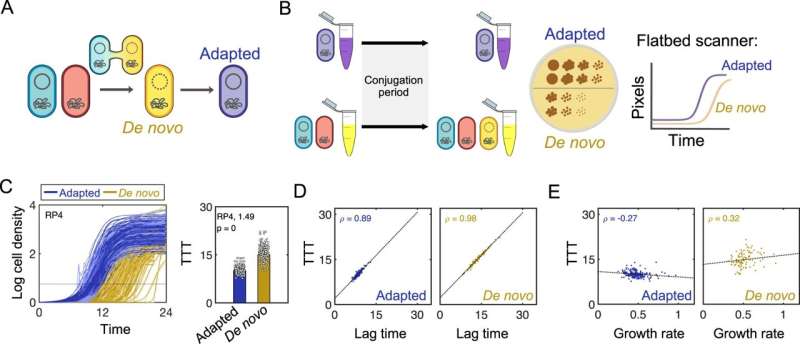
While common sense might suggest that plasmids that spread the easiest are the ones that allow bacteria to grow the fastest, a new study in Nature Communications, led by Allison Lopatkin, an assistant professor of chemical engineering at the University of Rochester, outlines the surprising evolutionary tradeoff between lag time and growth rate.
Fast acquisition of plasmids comes at a cost
"You would think that something that's able to grow faster will always do better, but we found that's not true because the acquisition costs manifest in a delay rather than a growth rate," says Lopatkin. "Taking a little bit longer to let that plasmid become established ultimately helped the gene spread faster."
Lopatkin and her team studied the growth rates of single colonies of bacteria immediately following plasmid acquisition. Across nearly 60 conditions covering diverse plasmids, selection environments, and clinical strains, they found that intermediate-cost plasmids outcompete both their low and high-cost counterparts.
The research shows plasmid costs are more complex than previously believed and is a step toward better understanding why certain types of pathogens are better at acquiring plasmids than others. If scientists can understand what controls the costs of acquiring a plasmid, they can potentially use that information to limit the spread of antibiotic-resistant genes.
"We see horizontal gene transfer as an engineering tool to control how genes can spread and help bacterial communities interact," says Lopatkin. "By understanding the individual parts, we hope not only to be able to fight things like antibiotic resistance, but also to use plasmids to deliver genes that can help natural bacteria degrade oil from oil spills. There are many applications microbiomes can be useful for."
Lopatkin and her lab plan to further investigate the underlying genetic and environmental conditions that successfully lead to horizontal gene transfer. Lopatkin's team will use computational modeling, bioinformatics, and mechanistic experiments to study the molecular factors favoring the formation of new strain-plasmid combinations.
"Ultimately, we hope to be able to predict high-risk gene transfer before it occurs, thereby allowing us to explore novel control and treatment strategies," says Lopatkin.
More information: Mehrose Ahmad et al, Tradeoff between lag time and growth rate drives the plasmid acquisition cost, Nature Communications (2023). DOI: 10.1038/s41467-023-38022-6
Journal information: Nature Communications
Provided by University of Rochester