This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Researchers study diversity and impact of broad host range plasmids in human gut
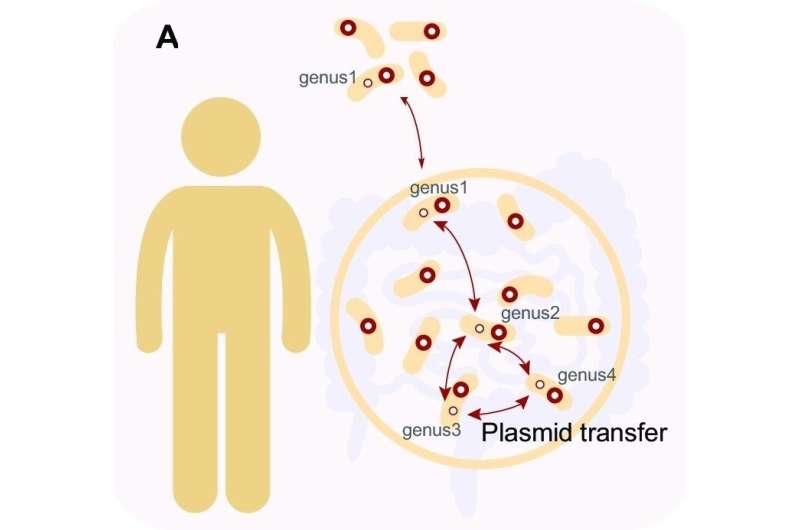
The human gut hosts a complex ecosystem comprising an intricate plasmidome with abundant undiscovered plasmids, driving horizontal gene transfer events.
Broad host range (BHR) plasmids carrying fitness genes transfer between bacteria across distantly phylogenetic taxa, facilitating the adaptation of their hosts to varying environments. However, accurate identification of BHR plasmids in the gut is still difficult, and their host range and prevalence remain unclear.
Recently, a research team led by Prof. Ma Yingfei from the Shenzhen Institute of Advanced Technology (SIAT) of the Chinese Academy of Sciences has identified and characterized plasmids harbored by human gut bacterial isolates from culturomics-based studies. The researchers have revealed their accurate host range, persistence in human gut, accessory genes, prevalence across various environments, and transmission and evolutionary trajectories in detail.
In the study, published in Nucleic Acids Research, researchers assembled 820 plasmid-like clusters (PLCs) with high-completeness genomes (comPLCs). "The majority of PLCs (81.1%) were classified to known replicon types, which suggested gut plasmids were highly diverse and novel," said Yang Lili, first author of the study.
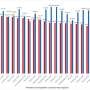
They found that 175 comPLCs had a broad host range across distinct bacterial genera, of which, 71 were detected in at least two human populations from four distant countries, and 13 were highly prevalent (>10%) in at least one human population. They observed that the broadest PLC (Clstr-417) could spread among the isolates collected from Chinese donors and American donors of 24 genera across five phyla. And some PLCs are so ubiquitous that can even be detected in different species and environmental sewage.
The evolutionary trajectory showed that the BHR plasmid spread between the phyla and the environment was extremely rapid, with all transmission times occurring 0-10 years ago and colonizing the individual gut multiple times. "We found that these BHR plasmids could carry many different adaptive genes, and individual PLCs even could carry up to seven antibiotic resistance genes," said Prof. Ma.
Prof. Ma is considering more possibilities for BHR plasmids. "BHR plasmids may also have great potential to edit the genome of commensal gut microbes and bacteriophages that suffer from lacking editing tools," he said.
More information: Lili Yang et al, Global transmission of broad-host-range plasmids derived from the human gut microbiome, Nucleic Acids Research (2023). DOI: 10.1093/nar/gkad498
Journal information: Nucleic Acids Research
Provided by Chinese Academy of Sciences