August 30, 2023 feature
This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
A DNA assembly kit to unlock the CRISPR-Cas9 potential for metabolic engineering
The clustered regularly interspaced short palindrome repeats (CRISPR) and Crispr-associated protein 9 (CRISPR/Cas9) is now a well-known, revolutionary method to engineer microbial cells.
A key advantage of CRISPR remains in the strain design to facilitate chromosomal integration to enable the assembly of marker-free DNA. These editing systems are highly beneficial; however, their assembly is not quite straightforward and may prevent its use and applications.
In a new report in Nature Communications Biology, Tigran V. Yuzbashev and a research team identified the limits of the existing Cas9 toolkits designed to make CRISPR techniques easier to access and implement. They discussed three different well-established methods and combined them to form a comprehensive toolkit for efficient metabolic engineering by using CRISPR/Cas9.
A single toolkit comprised of 147 plasmids to generate and characterize a library of 137 promoters to build a homogentisic acid in the lab.
Genome modifications with CRISPR/Cas9
The CRISPR/Cas9 system can render quick, precise and scarless genomic modifications to provide significant scope to design microbial strains for bioproduction. Metabolic engineering of yeasts for instance provide a fast-growing area in engineering biology for the sustained production of chemicals, fuels, foods, and pharmaceuticals.
Yeasts have a metabolic potential like eukaryotic cells and are therefore easier to engineer and cultivate at scale. As a result, bioengineers have designed and developed CRISPR systems for yeasts.
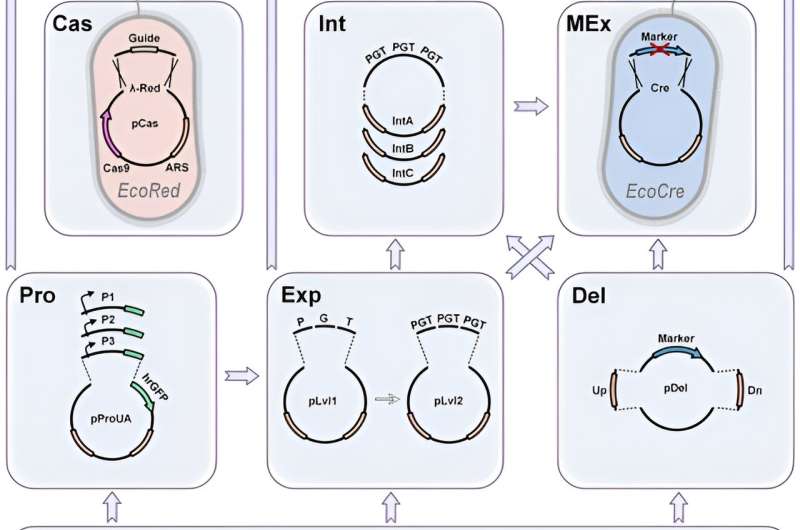
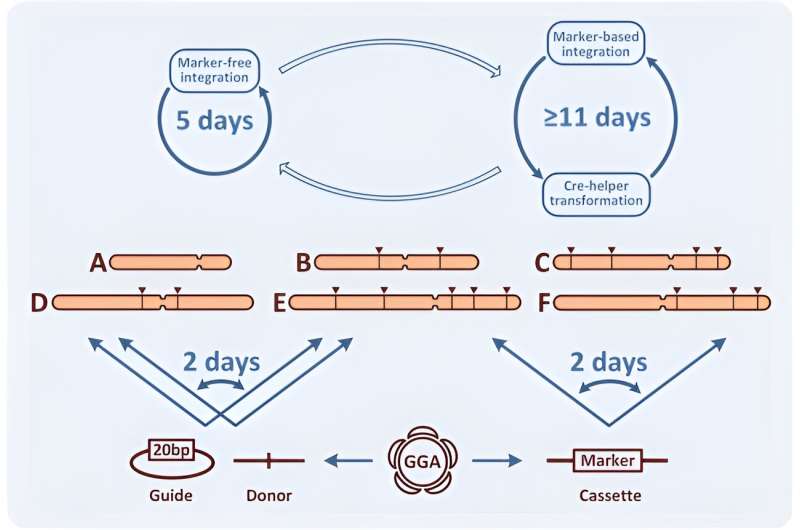
Due to its high efficiency, CRISPR allows marker-less genomic modifications. In this work, Yuzbashev ensured strain optimization and facilitated metabolic engineering projects by identifying three improvements of the CRISPR/Cas9 system for yeast engineering. The methods included: 1) the easy swap between marker and marker-less modifications, 2) the quick exchange of homology arms to determine different locations of integration, and 3) an easy method to clone gRNAs.
Marker-free integration
To enable CRISPR-based marker-free integration, the team chose a double strand break induced by Cas9, which had to be repaired to accomplish cell proliferation. The scientists made this possible by using a template or donor, integrated through homologous recombination or non-homologous end-joining (NHEJ)—without integration. The process of non-homologous end-joining is observed in most fungal species including baker's yeast Saccharomyces cerevisiae.
In species with a predominant NHEJ mechanism, the team enhanced homologous recombination by deleting the NHEJ genes. If a marker-free method did not succeed, the scientists subsequently aim to improve CRISPR-Cas9 assisted integration to easily revert to marker-based integration.
Donor DNA re-direction
The Cas9-assisted integration typically requires a donor template consisting of an integrated cassette flanked by two homology arms. The team theorized that the ideal integration of CRISPR/Cas9 should exchange homology arms on pre-assessed Cas9 donor constructs via a simplified Golden Gate assembly reaction.
Furthermore, promoters are a key element to any metabolic engineering project to redirect flux towards the products of interest. Yuzbashev et al. used the industrial yeast Yarrowia lipolytica to develop a metabolic engineering toolkit that combined gene editing and DNA assembly strategies for high efficiency and versatility.
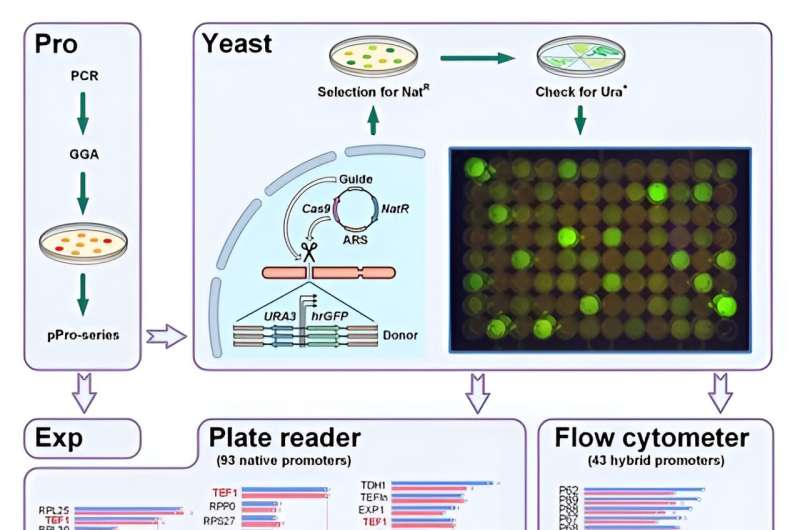
Modular architecture of the toolkit and metabolic pathway engineering
The scientists unlocked the full potential of CRISPR/Cas9 for metabolic engineering by developing a toolkit expanding on previously well-known Golden Gate assembly systems. They tested the screening system by generating several promoter libraries. Yuzbashev et al. chose Y. lipolytica ribosomal genes encoding proteins of large and small subunits. They identified a variety of promoters with diverse strengths to expand the number of promoters for the same organism.
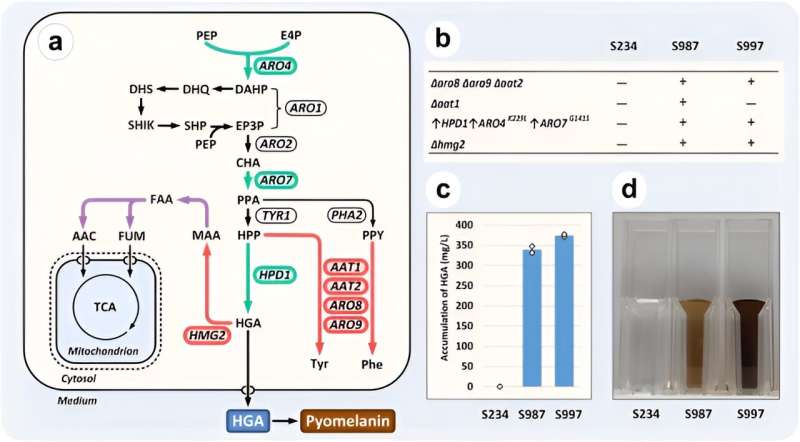
To prove the influence and use of the enhanced CRISPR/Cas9 method, the team created a Y. lipolytica via rational engineering to produce a homogentisic acid (HGA) . Typically under alkaline conditions HGA spontaneously undergoes oxidation to form self-polymerized pyomelanin; an excellent constituent of natural sunscreens and cosmetics.
Despite its high commercial potential, existing methods to produce the acid precursor and pyomelanin product relied on the biotransformation of expensive aromatic amino acids. To facilitate metabolic engineering, the team therefore first selected several genes that encoded theprecursor aromatic aminotransferases as engineering targets. They then selected three overexpression targets to enhance the de novo synthesis of the homogentisic acid in the model organism. Finally, they studied and inactivated the HGA degradation pathway; a path yet unknown to exist in Y. lipolytica.
Outlook
In this way, Tigran V. Yuzbashev and colleagues showed the dependence of metabolic engineering of living organisms on efficient DNA manipulation methods. This work presents an example of an enhanced molecular toolkit designed for CRISPR/Cas9-based metabolic engineering.
The scientists proved the functionality of the platform for both rapid strain construction and the characterization of a large library of promoters. They anticipate for this toolkit to have broader applications in strain engineering and in industry. The team envision for the Y. lipolytica model developed in this work to have overarching applications in other fields of biological engineering as well.
More information: Tigran V. Yuzbashev et al, A DNA assembly toolkit to unlock the CRISPR/Cas9 potential for metabolic engineering, Communications Biology (2023). DOI: 10.1038/s42003-023-05202-5
Guri Giaever et al, Functional profiling of the Saccharomyces cerevisiae genome, Nature (2002). DOI: 10.1038/nature00935
Journal information: Nature , Communications Biology
© 2023 Science X Network