February 14, 2023 report
This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Modified CRISPR/Cas9 gene editing system used to learn more about the evolution of giant viruses
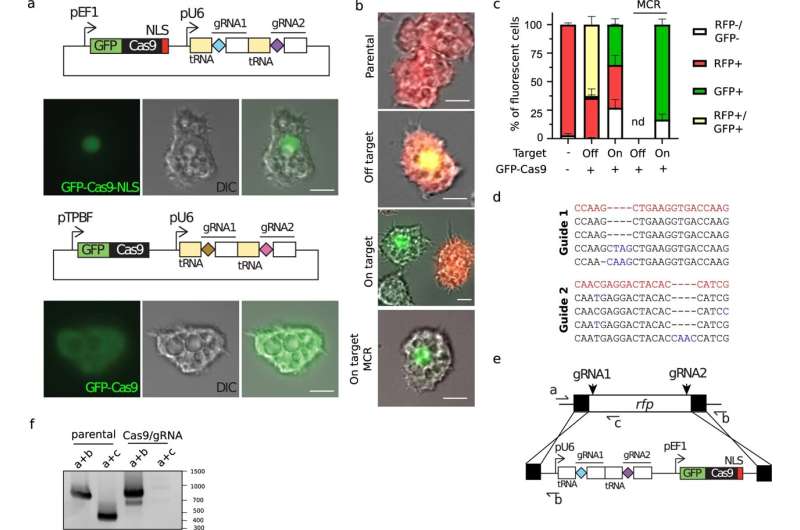
A team of virologists at Aix–Marseille University, has found evidence that suggests the giant virus Pandoravirus neocaledonia evolved from smaller and simpler viruses. In their study, published in the journal Nature Communications, the group used a modified version of the CRISPR/Cas9 gene editing system to learn more about the evolutionary history of the giant virus, and perhaps others like it.
Prior research has shown that most viruses are much smaller than bacteria. But a few are so large that biologists refer to them as giant viruses. Somewhat perplexed by their existence, evolutionary biologists have long debated how such strange viruses might have come to exist.
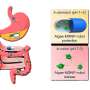
Currently, there are two main theories: The first is that they evolved as a mix of several smaller viruses. The second is that they devolved from larger, more sophisticated organisms. In this new effort, the team in France took a new approach to solving the mystery—using CRISPR/Cas9 to identify what are known as essential genes in the viruses' genome (those that are needed to reproduce).
Prior research has shown that a good way to discover essential genes is to remove genes one at a time until the organism is no longer able to reproduce. That is where CRISPR/Cas9 came in—it allows for knocking out whatever genes are desired. But Pandoravirus neocaledonia presented a problem—it has 25 copies of each of its chromosomes, and CRISPR/Cas9 is only able to knock out one gene at a time. To overcome this problem, the team modified the gene editing system to generate a chain reaction—whenever a gene was cut, another cut would be instigated along the chain until all of the copies were removed.
After running their chain-reaction gene editing on Pandoravirus neocaledonia until the virus was no longer able to reproduce, they found that the genes needed for reproduction were located on just one end of the genome, somewhat apart from other, less essential genes—evidence, the team suggests, that the virus evolved from several smaller viruses. They suggest that it is likely that other giant viruses evolved in similar ways.
More information: Hugo Bisio et al, Evolution of giant pandoravirus revealed by CRISPR/Cas9, Nature Communications (2023). DOI: 10.1038/s41467-023-36145-4
Journal information: Nature Communications
© 2023 Science X Network