Simultaneous mapping of several epigenetic landmarks in a single cell
Researchers at Karolinska Institutet and Stockholm University have developed a new technology allowing simultaneous probing of several different histone marks in one individual cell and in thousands of cells at the same time. This new method allows researchers to investigate in much greater detail how cells in the mouse brain acquire unique properties and specialize. The study is published in the journal Nature Biotechnology.
"This technique is a steppingstone to understand how cells can transition between different states," says Marek Bartosovic, previously post-doctoral fellow at the Department of Medical Biochemistry and Biophysics, Karolinska Institutet, and now research group leader at Stockholm University.
Cells in our bodies can be very different, but they inherit in their genome common genetic information in the form of DNA. To acquire different properties and specializations, cells read and interpret this common genome in different ways. One such mechanism of reading DNA involves proteins called histones, which are bound to the genome. Histones exhibit variable surface landmarks, which carry multiple layers of epigenetic information. These landmark histone modifications are present in different patterns in the genome of different cell types. This allows each cell to interpret the genomic information in a different manner, and then to acquire different properties.
Nano CUT&Tag
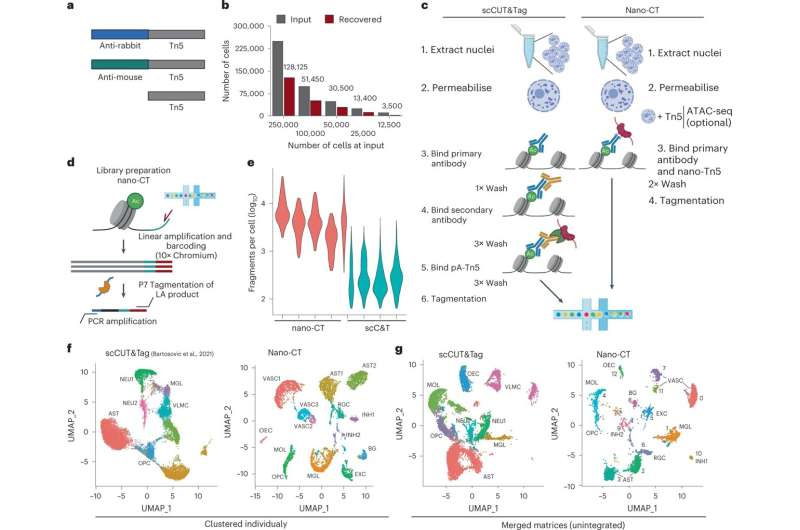
Recently, several research teams including the team at Karolinska Institutet, developed methods such as single cell CUT&Tag, allowing to look at individual histone modifications in a single cell level and at a large scale.
Now the Karolinska Institutet team has made a further leap by developing a novel technology, Nano CUT&Tag. This technology is based on a new class of molecules called nanobodies. Nanobodies are small proteins that work just like antibodies in recognizing other proteins with high specifity but are smaller and can easily be fused with other proteins. The use of different nanobodies fused with an enzyme called Tn5 transposase allowed the probing of different histone modifications at the same time in the same cell. Nano CUT&Tag gives unique insights how a cell can, via adjusting multiple layers of epigenetic information simultaneously, interpret the genome to specialize and acquire an unique identity.
"Nano CUT&Tag, or Nano-CT, as we call it, allowed us to dissect in very much detail how progenitor cells in the brain become specialized into oligodendrocytes, a cell type which is target of an auto-immune attack in multiple sclerosis," Gonçalo Castelo-Branco, Professor of Glial Cell Biology at Karolinska Institutet, says. "These new mechanistic insights can give us clues on how to stimulate the recovery of the oligodendrocyte population in the context of disease."
"We now want to further develop Nano-CT, by increasing the range of epigenetic landmarks it can probe," says Marek Bartosovic, who has recently started his research group at Stockholm University, where he will focus on the development of single cell epigenomic technologies, in the context of early human brain development.
More information: Marek Bartosovic et al, Multimodal chromatin profiling using nanobody-based single-cell CUT&Tag, Nature Biotechnology (2022). DOI: 10.1038/s41587-022-01535-4
Journal information: Nature Biotechnology
Provided by Karolinska Institutet