New algorithm uncovers secrets of cell factories
Drug molecules and biofuels can be made to order by living cell factories, where biological enzymes do the job. Now, researchers at Chalmers University of Technology have developed a computer model that can predict how fast enzymes work, making it possible to find the most efficient living factories, as well as to study complex diseases. Their results are published in Nature Catalysis.
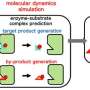
"To study every natural enzyme with experiments in a laboratory would be impossible, they are simply too many. But with our algorithm, we can predict which enzymes are most promising just by looking at the sequence of amino acids they are made up of," says Eduard Kerkhoven, researcher in systems biology at Chalmers University of Technology and the study's lead author.
Only the most promising enzymes need to be tested
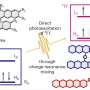
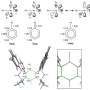
The enzyme turnover number or kcat value, describes how fast and efficient an enzyme works and is essential for understanding a cell's metabolism. In the new study, Chalmers researchers have developed a computer model that can quickly calculate the kcat value. The only information needed is the order of the amino acids that build up the enzyme—something that is often widely available in open databases. After the model makes a first selection, only the most promising enzymes need to be tested in the lab.
Given the number of naturally occurring enzymes, the researchers believe that the new calculation model may be of great importance.
"We see many possible biotechnological applications. As an example, biofuels can be produced when enzymes break down biomass in a sustainable manufacturing process. The algorithm can also be used to study diseases in the metabolism, where mutations can lead to defects in how enzymes in the human body work," says Eduard Kerkhoven.
More knowledge on enzyme production
More possible applications are more efficient production of products made from natural organisms, as opposed to industrial processes. Penicillin extracted from a mold is one such example, as well as the cancer drug taxol from yew and the sweetener stevia. They are typically produced in low amounts by natural organisms.
"The development and manufacture of new natural products can be greatly helped by knowledge of which enzymes can be used," says Eduard Kerkhoven.
The calculation model can also point out the changes in kcat value that occur if enzymes mutate, and identify unwanted amino acids that can have a major impact on an enzyme's efficiency. The model can also predict whether the enzymes produce more than one "product."
"We can reveal if the enzymes have any 'moonlighting' activities and produce metabolites that are not desirable. It is useful in industries where you often want to manufacture a single pure product."
The researchers tested their model by using 3 million kcat values to simulate metabolism in more than 300 types of yeasts. They created computer models of how fast the yeasts could grow or produce certain products, like ethanol. When compared with measured, pre-existing knowledge, the researchers concluded that models with predicted kcat values could accurately simulate metabolism.
More information: Feiran Li et al, Deep learning-based kcat prediction enables improved enzyme-constrained model reconstruction, Nature Catalysis (2022). DOI: 10.1038/s41929-022-00798-z
Journal information: Nature Catalysis
Provided by Chalmers University of Technology