New yeast model can improve protein production
Microorganisms, such as baker's yeast, can be used as cell factories to produce different chemicals and proteins, such as commonly used pharmaceuticals as insulin. By modifying the cell factories researchers are trying to increase the yield and speed of the production processes. In a new study published in Nature Communications, researchers in systems biology at Chalmers provide us with a new yeast model focusing on one of the limiting steps of the cell factory production—the secretory pathway.
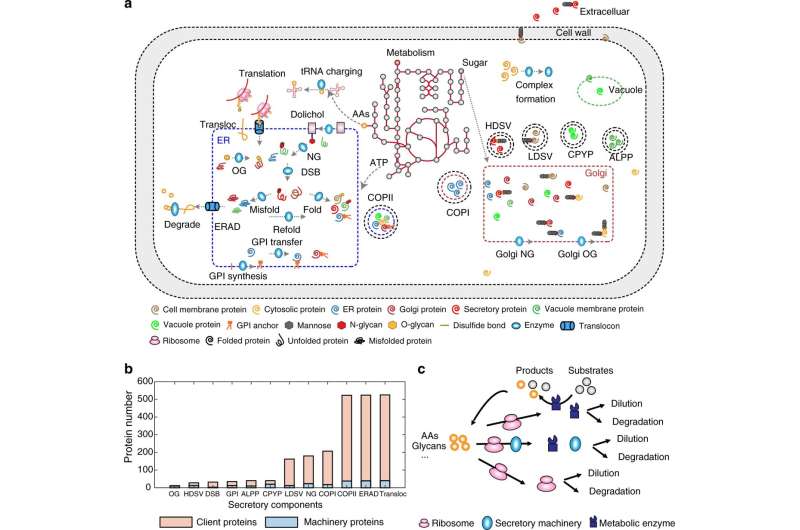
The secretory pathway in eukaryotic cells consists of several different organelles that involve transportation and different modifications of proteins. This very complex pathway is—and has been historically—a target for improving recombinant protein expression in the cells in order to develop efficient cell factories. But due to its natural complexity, there has so far been a lack of tools to systematically optimize this pathway to produce any recombinant protein.
Advanced model can lead to better cell factory production
In their recently published study, the researchers present a new advanced genome-scale protein secretory model of baker's yeast, Saccharomyces cerevisiae, called pcSecYeast.
"We aimed to systematically understand protein secretion in yeast to better design the yeast cell factory for recombinant pharmaceutical and industrial production of proteins such as insulin and α-amylase, an enzyme that converts starch into sugars," says Feiran Li, postdoc in systems and synthetic biology at Chalmers and first author of the study.
Model predicts engineering targets
The model is the first complex and advanced protein secretory model containing the detailed processes of how the proteins are synthesized and modified to their mature form in the cell. This model enables multiple types of simulations, including the competition between those recombinant and native secretory proteins for the limited resources.
"We predicted the engineering targets for eight recombinant proteins produced in yeast and experimentally validated several predicted targets for α-amylase. For the first time, a model can systematically predict the engineering targets in both the metabolic and protein secretory part," says Feiran Li.
Rational cell factory strain design
By identifying engineering targets (such as gene amplification targets) the model can be used for rational cell factory strain design, for industrial or pharmaceutical protein production.
"The model also facilitates in silico testing of various hypotheses for protein expression and secretion, which will boost the fundamental understanding of the complex secretory pathway," says Feiran Li.
"We also tested several hypotheses and identified that the so called retro-translocation capacity could prevent degradation of excessive misfolded protein. This can lead to the accumulation of those proteins, which is related to human diseases such as Alzheimer's and Parkinson's."
More information: Feiran Li et al, Improving recombinant protein production by yeast through genome-scale modeling using proteome constraints, Nature Communications (2022). DOI: 10.1038/s41467-022-30689-7
Journal information: Nature Communications
Provided by Chalmers University of Technology