A study uncovers the 'grammar' behind human gene regulation
A research group at the University of Helsinki has discovered the logic that controls gene regulation in human cells. In the future, this new knowledge could be used for investigating cancers and other genetic diseases.
Gene regulation is an important process that controls the activity of genes in cells. Incorrect gene regulation can contribute to the onset of many diseases, including cancer.
The DNA of the human genome contains genes that code for proteins, which in turn give muscle cells their strength and brain cells their ability to process information. DNA also contains gene regulatory elements that determine when and where genes are expressed, ensuring muscle genes are expressed in muscles and brain genes in the brain.
However, the regulatory code that determines gene activity remains poorly understood. Even though the human genome comprises almost 3 billion base pairs, the genomic sequence alone is too short for learning the gene regulatory code. The problem is similar to that faced by a linguist who tries to understand a forgotten language on the basis of a few short texts.
A research group of professor Jussi Taipale that belongs to the Academy of Finland's Centre of Excellence in Tumour Genetics Research has now found a way around this problem to solve the regulatory code.
The new study was recently published in the Nature Genetics journal.
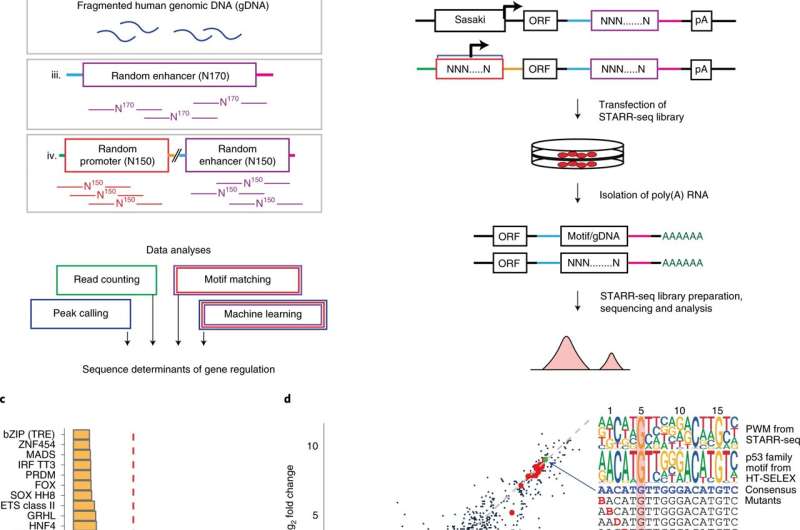
"We measured the gene regulatory activity from a collection of DNA sequences that together are 100 times larger than the entire human genome," says Academy of Finland Research Fellow Biswajyoti Sahu, the first author of the study.
"Instead of using the natural genomic sequence, we introduced random synthetic DNA sequences to human cells. Then, the cells themselves were allowed to read the new DNA and highlight for us the sequences that function as active regulatory elements," Sahu adds, describing the innovative approach.
Researchers identify the key atomic unit of gene expression
The researchers produced their extensive dataset using a technique known as massively parallel reporter assay, where the regulatory activity of millions of DNA sequences can be simultaneously studied in one large-scale assay. The data were analyzed using artificial intelligence tools.
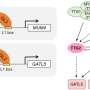
Gene expression is regulated by proteins, known as transcription factors, that bind the DNA. The researchers found that the very short DNA sequences to which these factors bind constitute the key atomic unit of gene expression. Individual transcription factors contribute to gene regulation in an additive manner. In other words, each factor increases regulatory activity independently without specific interactions with other factors. In addition, transcription factors may have several parallel functions in the gene regulatory process, such as enhancing the rate of gene expression or defining the genomic location where the transcription starts.
"The binding motifs of transcription factors can be thought to be like words that together define the cellular gene regulatory code," professor Jussi Taipale explains.
The researchers found that the grammar for the code is relatively weak, and that most words can be placed in almost any order without changing their meaning.
"However, in some cases analogous to compound words, the grammar is strong, and specific combinations of factors need to bind in a certain order to activate gene expression," says Taipale.
Only a handful of highly active transcription factors in cells
The researchers compared three different human cell types: colon and liver cancer cells as well as normal cells originating from the retina. They found that only a handful of transcription factors are highly active in cells. Furthermore, most transcription factor activities are similar regardless of cell type.
The results revealed that the gene regulatory elements in the human cells can be classified into different types based on the chromatin context they are located in—either in closed chromatin regions with densely packed DNA, or in a more open chromatin environment where the DNA is not as tightly packed around histone proteins.
Traditionally, active regulatory elements have been thought to be located within open chromatin regions where DNA is easily accessible to transcription factors. Thus, the discovery of active regulatory elements that function within closed chromatin regions is one of the central new observations of the study. In addition, the researchers identified regulatory elements that are dependent on chromatin. These elements are active at their normal sites in the genome, but their activity drops considerably if they are removed from their original location and transferred close to another gene.
More information: Biswajyoti Sahu et al, Sequence determinants of human gene regulatory elements, Nature Genetics (2022). DOI: 10.1038/s41588-021-01009-4
Journal information: Nature Genetics
Provided by University of Helsinki