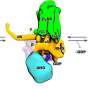
Proteins engineered to form honeycomb structures can block uptake of receptors from the surface of cells

A new class of protein material that interacts with living cells without being absorbed by them can influence cell signaling, a new study shows. The material does this by binding and sequestering cell surface receptors.
The discovery could have far-reaching implications for stem cell research and enable the development of new materials designed to modulate the behavior of living systems.
The research, reported in the January 6 edition of Nature, was led by the Baker lab at the University of Washington School of Medicine and the Derivery lab at the Medical Research Council Laboratory of Molecular Biology in Cambridge, U.K. Their paper is titled, Design of Biologically Active Binary Protein 2-D Materials.
Cells interact with their environment via receptors at their surface. These receptors can bind to hormones, neurotransmitters, drugs, and toxins. When such molecules bind to a receptor, this triggers a response inside the cell, a process known as signaling.
But for the cell, it is important that this response be transient, to still be responsive to the signal later on. To achieve this, cells will commonly terminate signaling by absorbing both an activated receptor and the molecule that stimulated it, thereby targeting both for destruction inside the cell.
"This tendency of cells to internalize receptors likely lowers the efficiency of immunotherapies," said Emmanuel Derivery, assistant professor at the MRC Laboratory of Molecular Biology. "Indeed, when antibody drugs bind their target receptors and then become internalized and degraded, more antibody must always be injected."
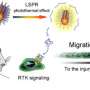
To create a way around this, Baker lab postdoctoral scholar Ariel Ben-Sasson designed new proteins that assemble into large, flat patches. This molecular scaffolding was then further engineered to contain signaling molecules.
Graduate student Joseph Watson of the Derivery lab showed that such protein materials could latch onto cells, activate surface receptors, and resist being absorbed by the cell for hours or even days.
"This work paves the way towards a synthetic cell biology, where a new generation of multi-protein materials can be designed to control the complex behavior of cells," said David Baker, professor of biochemistry at the UW School of Medicine and director of the UW Medicine Institute for Protein Design.
By swapping out which cell surface receptors were targeted by the patch, the researchers showed that different cell types could be activated.
"We now have a tool that can interact with any type of cells in a very specific way," said Ben-Sasson. "This is what is exciting about protein engineering: it opens fields that people may not expect."
According to co-author Hannele Ruohola-Baker, professor of biochemistry at the UW School of Medicine and associate director of the UW Medicine Institute for Stem Cell and Regenerative Medicine, versions of these new materials could eventually help physicians alleviate the dangers of sepsis by controlling the inflammatory response to infection.
They might even enable entirely new ways to treat COVID-19, heart disease, and diabetes, and perhaps mitigate the downstream effects of strokes, including Alzheimer's disease.
"This breakthrough helps pave the way for the use of synthetic cell biology in regenerative medicine," said Ruohola-Baker.
More information: Design of biologically active binary protein 2D materials, Nature (2021). DOI: 10.1038/s41586-020-03120-8 , www.nature.com/articles/s41586-020-03120-8
Journal information: Nature
Provided by University of Washington