New method accelerates development of protein therapeutics
Glycosylation—the attachment of sugars to proteins—plays a critical role in both cellular function and in the development of therapeutics, like vaccines.
But because researchers have used mammalian cells to create the biosynthetic pathways (sets of enzyme catalysts) to build and study these sugar structures, the process has historically taken a long time and has required specialized laboratory equipment.
Northwestern Engineering researchers have now developed a quick, cell-free system to build and study these pathways. Called GlycoPRIME, the system could lead to faster development of therapeutics and a new, modular way to make medicines on demand in resource-limited settings.
"This is an exciting new method that accelerates the design and engineering of potential medicines and vaccines using glycosylation," said Michael Jewett, the Charles Deering McCormick Professor of Teaching Excellence, professor of chemical and biological engineering, and director of Northwestern's Center for Synthetic Biology, who led the research. "It's opening a new book to engineer proteins not seen in nature with specific applications."
The results were published November 27 in the journal Nature Communications. Milan Mrksich, the Henry Wade Rogers Professor of Biomedical Engineering, Chemistry, and Cell and Molecular Biology at Northwestern's McCormick School of Engineering, is a co-author on the paper. He is also the interim vice president for research at Northwestern.
A new approach to discovering glycosylation pathways
Glycosylation is important in the development of protein medicines, which include everything from anti-cancer drugs like Herceptin to flu and tetanus vaccines. Sugar structures allow these proteins to remain stable while enabling them to perform tasks, like attack a cancer cell or retrain the immune system.
Developing these medicines has generally required many years and a laboratory with highly specialized equipment to grow mammalian cells. These mammalian cells naturally produce glycosylated proteins, but are slow-growing and can be difficult to engineer, limiting the number and diversity of glycosylation structures that can be built and tested.
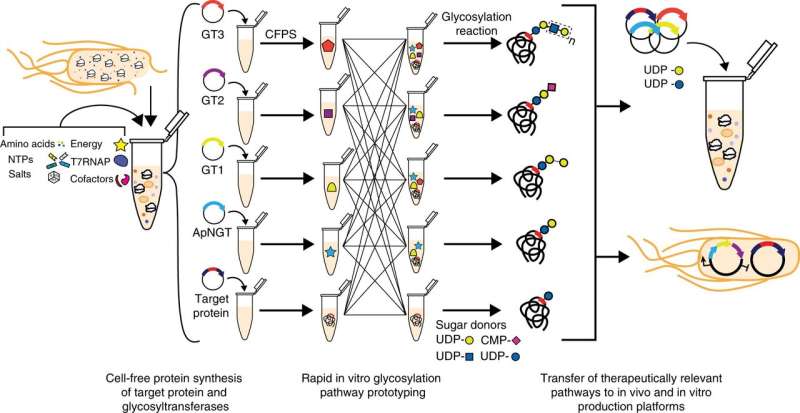
Jewett's lab has developed cell-free systems that create enzymes needed to create certain proteins, but up until now, these processes could not create glycosylated products without the need to reengineer living cells.
Weston Kightlinger, a Ph.D. student in the Jewett lab, developed a new approach to build, test, and assess sets of enzymes that can modularly build sugars for protein therapeutics. The process involves producing and then mixing and matching enzymes in test tubes to discover and understand which enzymes are needed to build desired sugar structures.
Kightlinger likens the process to building a flying machine. Instead of building a bird, which is complex and difficult to create, humans build airplanes, which are much simpler than birds but can be engineered to fly just the same. "We are pipetting liquids instead of rebuilding cells," he said. "Doing this, you can go from DNA to a glycoprotein in a matter of hours rather than weeks."
In just a few months, Kightlinger used the system to construct 37 pathways, creating 23 unique sugar structures, 18 of which have never been synthesized on proteins.
"We can now mix and match to build biosynthetic pathways an order of magnitude faster than what has been conventionally done," Jewett said. "The dream is to create new-to-nature glycoproteins that could be made on-demand to develop new types of protein medicines."
A new method for developing therapeutics and vaccines
The researchers used this process to develop a protein vaccine candidate modified with a sugar structure that could trigger the immune system, as well as a therapeutic antibody fragment with a sugar that can stabilize proteins as they circulate in the body. Future works will use other pathways developed in this paper to create glycosylated protein vaccines and therapeutics that can target certain areas within the body. The system could also be used to provide modular, on-demand biomanufacturing platforms that provide medicines or vaccines in resource-limited settings.
"Because this system is so robust and simple, it can teach us more about how these sugar structures actually work," Kightlinger said. "That will allow us to optimize them to better understand which of these structures to pursue."
More information: Weston Kightlinger et al, A cell-free biosynthesis platform for modular construction of protein glycosylation pathways, Nature Communications (2019). DOI: 10.1038/s41467-019-12024-9
Journal information: Nature Communications
Provided by Northwestern University