Tale of two trees: New web tool estimates gene trees with ease
Gene trees, much like family trees, trace the lineage of a particular gene from its deep ancestral roots to its still-growing branches. By comparing gene trees to species trees, which map the evolutionary history of species, scientists can learn which species have which genes, what new functions those genes gained over time, and which functions they may have lost. Now, scientists at the Okinawa Institute for Science and Technology Graduate University (OIST) have unveiled a new tool to perform these analyses quickly and without computational headaches.
The free, web-based tool ORTHOSCOPE consults well-established species trees and about 250 genomic datasets to estimate gene trees and identify orthogroups—sets of genes descended from a single gene in the last common ancestor of a select group of species. Start to finish, the analysis takes only a few minutes. The researchers described the tool and multiple case studies validating its efficacy in a new paper, published December 4, 2018, in Molecular Biology and Evolution.
The speedy software allows researchers to identify if a gene is present in a species' genome and how many copies there are. Most importantly, it makes it simple to rapidly infer the function of that gene, as well as the functions of its ancestors.
"We need to think about species evolution when we think about gene function," said Jun Inoue, first author of the study and a staff scientist in the Marine Genomics Unit, led by Prof. Noriyuki Satoh. Gene trees require significant time, effort and data to construct manually, he said, so in the past, many studies have investigated gene function without this contextualizing information. By estimating gene trees automatically, Inoue's new software could greatly improve studies of gene function in bilateral animals—including humans.
"This software makes it possible to compare the phylogenetic relationship of different genes," Inoue said. "I hope the tool is used in medical research—it makes a big difference."
Streamlining a once difficult process
Prior to the launch of ORTHOSCOPE, collections of genomic data were scattered far and wide. The ability to build accurate gene trees relies on having access to adequate genomic data, but it takes time and effort to gather data from every corner of the web. To ease the process, Inoue and Satoh compiled data from the NCBI and Ensembl gene banks, along with a large database already built by the Marine Genomics Unit.
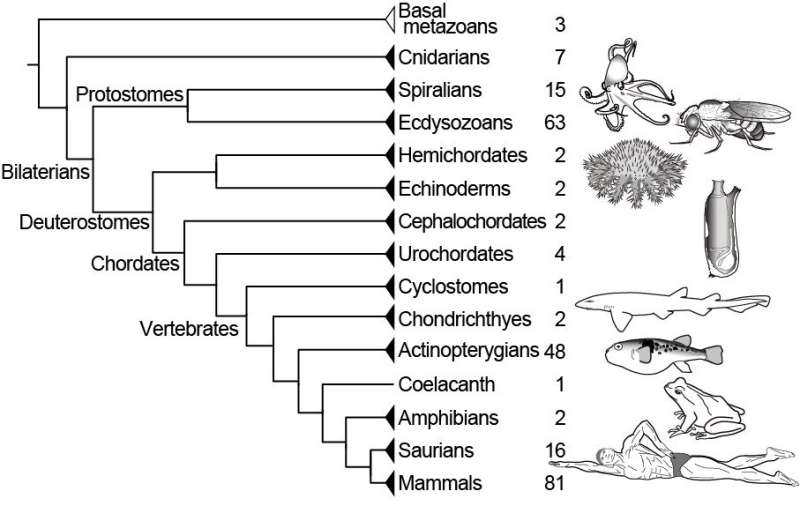
ORTHOSCOPE users start an analysis by inputting the coding sequences of protein-coding genes they're interested in. They then select one of four groups of species—namely, Protostomia, Deuterostomia, Vertebrata or Actinopterygii—to focus their search. They can refine their query further by selecting specific species to sample. Given sequence data, ORTHOSCOPE automatically estimates a new gene tree and delivers results within minutes. Users can rearrange the resulting tree based on a default species tree, provided by the software, or on data they provide themselves.
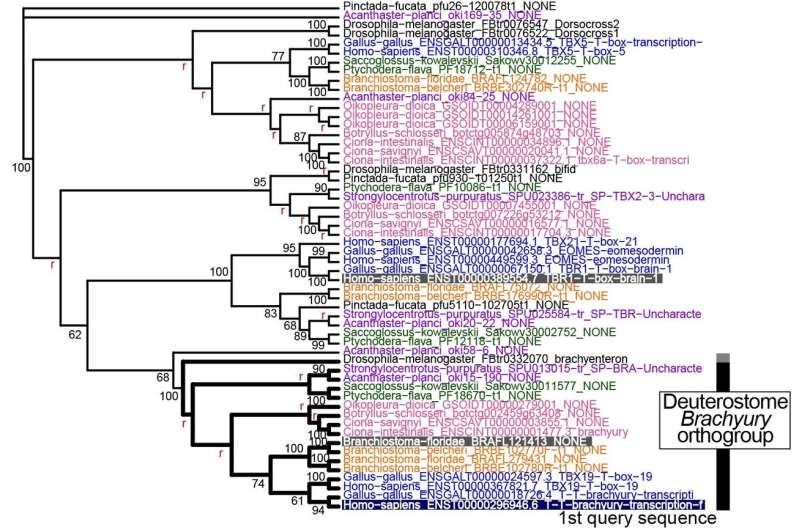
To test their new tool, Inoue and Satoh ran a few case studies of their own. For example, the researchers used ORTHOSCOPE to determine how many copies of the Brachyury gene, which is crucial to the development of the notochord, are present in different deuterostome species. The software confirmed results the researchers had collected manually in a previous study, but did so in significantly less time.
In another case study, the scientists were able to identify genes that evolved as a result of whole genome duplication, a key event in vertebrate evolutionary history. Whole genome duplication essentially quadrupled the size of the ancestral vertebrate genome, opening the door for more random mutations and the introduction of novel gene functions.
These case studies demonstrate that, with ORTHOSCOPE, researchers can go beyond comparing genes one by one and learn how they evolved and which species they impacted along the way.
"There is no other good method to estimate or infer gene function—this software does it automatically, and fast," said Inoue. "We can now know the entire history of a gene."
More information: Jun Inoue et al, ORTHOSCOPE: an automatic web tool for phylogenetically inferring bilaterian orthogroups with user-selected taxa, Molecular Biology and Evolution (2018). DOI: 10.1093/molbev/msy226
Journal information: Molecular Biology and Evolution
Provided by Okinawa Institute of Science and Technology