Bringing out the best in X-ray crystallography data
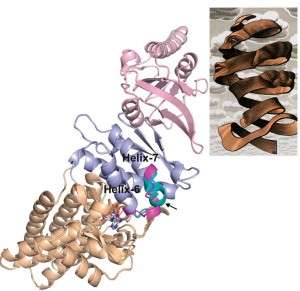
(Phys.org) —"Function follows form" might have been written to describe proteins, as the M. C. Escher-esque folds and twists of nature's workhorse biomolecules enables each to carry out its specific responsibilities. Technology's workhorse for determining protein structures is X-ray protein crystallography, in which a beam of x-rays sent through a crystallized protein is scattered by the protein's atoms, creating a diffraction pattern of dots that can be reconstructed by computer into a 3D model
While synchrotron radiation facilities, such as Berkeley Lab's Advanced Light Source, have been a boon to the field of protein crystallography, providing increasingly higher resolution structures over increasingly shorter time-spans, the technology is still a challenge. For some molecules, especially large molecular complexes, it is often only possible to obtain low-resolution experimental data, which means models are difficult to make and must be manually refined using computer modeling.
"Refinement of protein and other biomolecular structural models against low-resolution crystallographic data has been limited by the ability of current methods to converge on a structure with realistic geometry," says Paul Adams, a bioengineer with Berkeley Lab's Physical Biosciences Division and leading authority on x-ray crystallography, who, starting in 2000, has been leading the development of a highly successful software program called PHENIX (Python-based Hierarchical ENvironment for Integrated Xtallography) that automates crystallography data analysis.
Now, Adams and a team that included Nathaniel Echols in his research group, and Frank DiMaio with the research group of David Baker at the University of Washington, have developed a new method for refining crystallographic data that combines aspects of PHENIX with aspects of Rosetta, the most widely used software for the prediction and design of the three-dimensional structure of proteins and other large biomolecules.
The Rosetta program, which was originally developed by Baker and his research group, utilizes a detailed all-atom force field plus a diverse set of search procedures for the creation of its 3D models. PHENIX assembles 3D models atom-by-atom through the extraction of the best data from X-ray measurements. One of the most important components of PHENIX is "phenix.refine," a program for improving these models against the X-ray data using maximum likelihood methods. It was this feature that was combined with Rosetta.
"Our new method integrates the Rosetta and PHENIX programs directly in a flexible framework that allows it to be adapted to a wide variety of different scenarios," says Echols. "The main advantage of our method is that it can aggressively optimize models to fit the data and also present realistic geometry. In general, it has been difficult to come up with methods that handle both of these demands. As a result, crystallographers have either spent a lot of time fixing errors, or the published structures end up being of poor quality."
More information: "Improved low-resolution crystallographic refinement with Phenix and Rosetta," Nature Methods, 2013.
Journal information: Nature Methods
Provided by Lawrence Berkeley National Laboratory