This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Researchers identify over 2,000 potential toxins using machine learning
In a novel study, researchers have unveiled new secrets about a fascinating bacterial weapon system that acts like a microscopic syringe. The research paper, titled "Identification of novel toxins associated with the extracellular contractile injection system using machine learning" is published in Molecular Systems Biology
Led by Dr. Asaf Levy from the Hebrew University and collaborators from the Hebrew University and from the University of Illinois Urbana-Champaine, the team has made significant strides in understanding the extracellular contractile injection system (eCIS), a unique mechanism used by bacteria and archaea to inject toxins into other organisms.
Cracking the bacterial code with AI
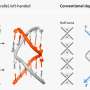
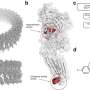
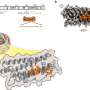
The eCIS is a 100-nanometer long weapon that evolved from viruses that previously attacked microbes (phages). During evolution, these viruses lost their ability to infect microbes and turned into syringes that inject toxins into different organisms, such as insects.
Previously, the Levy group identified eCIS as a weapon carried by more than 1,000 microbial species. Interestingly, these microbes rarely attack humans, and the eCIS role in nature remains mostly unknown. However, we know that it loads and injects protein toxins.
The specific proteins injected by eCIS and their functions have long remained a mystery. Before the study we knew about ~20 toxins that eCIS can load and inject.
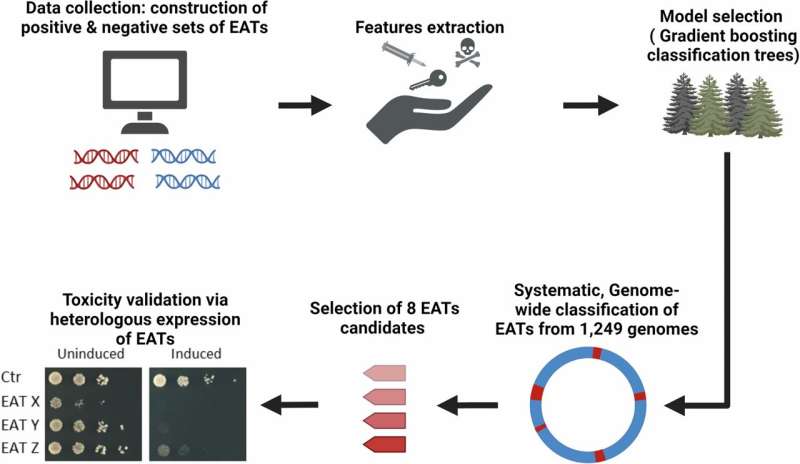
To solve this biological puzzle, the research team developed an innovative machine learning tool that combines genetic and biochemical data of different genes and proteins to accurately identify these elusive toxins. The project resulted in identification of over 2,000 potential toxin proteins.
"Our discovery not only sheds light on how microbes interact with their hosts and maybe with each other, but also demonstrates the power of machine learning in uncovering new gene functions," explains Dr. Levy. "This could open up new avenues for developing antimicrobial treatments or novel biotechnological tools."
New toxins with enzymatic activities against different molecules
Using AI technology, the researchers analyzed 950 microbial genomes and identified an impressive 2,194 potential toxins. Among these, four new toxins (named EAT14-17) were experimentally validated by demonstrating that they can inhibit growth of bacteria or yeast cells.
Remarkably, one of these toxins, EAT14, was found to inhibit cell signaling in human cells, showcasing its potential impact on human health. The group showed that the new toxins likely act as enzymes that damage the target cells by targeting proteins, DNA or a molecule that is critical to energy metabolism. Moreover, the group was able to decipher the protein sequence code that allows loading of toxins into the eCIS syringe.
Recently, it was demonstrated that eCIS can be used as a programmable syringe that can be engineered for injection into various cell types, including brain cells. The new findings from the current paper leverage this ability by providing thousands of toxins that are naturally injected by eCIS and the code that facilitates their loading into the eCIS syringe. The code can be transferred into other proteins of interest.
From microscopic battles to medical breakthroughs
The study's findings could have far-reaching applications in medicine, agriculture, and biotechnology. The newly identified toxins might be used to develop new antibiotics or pesticides, efficient enzyme for different industries, or to engineer microbes that can target specific pathogens.
This research highlights the incredible potential of combining biology with artificial intelligence to solve complex problems that could ultimately benefit human health.
"We're essentially deciphering the weapons that bacteria evolved and keep evolving to compete over resources in nature," adds Dr. Levy. "Microbes are creative inventors and it is fulfilling to be part of a group that discovers these amazing and surprising inventions."
The study was led by two students: Aleks Danov and Inbal Pollin from the department of Plant Pathology and Microbiology, the Institute of Environmental Sciences.
More information: Aleks Danov et al, Identification of novel toxins associated with the extracellular contractile injection system using machine learning, Molecular Systems Biology (2024). DOI: 10.1038/s44320-024-00053-6
Journal information: Molecular Systems Biology
Provided by Hebrew University of Jerusalem