This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
proofread
Study highlights potential for genetic manipulation in cucumber breeding
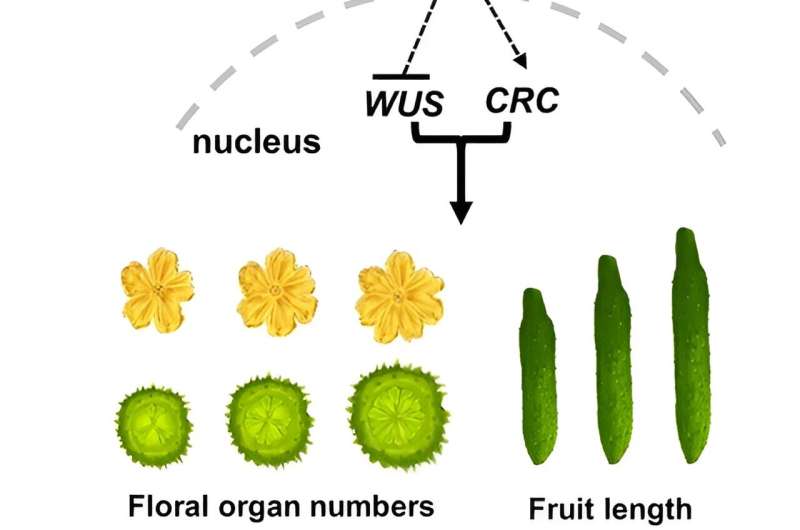
The CLAVATA (CLV) signaling pathway is crucial for controlling flower and fruit development by regulating the shoot apical meristem (SAM) size. Despite its significance, the downstream signaling components in crops remain largely unknown. Understanding these pathways is essential for advancing crop breeding techniques to enhance yield and quality.
There is a pressing need to investigate the genetic interactions within the CLV signaling pathway to uncover new regulatory mechanisms and improve agricultural practices.
A team from China Agricultural University, Beijing, investigated how the Gα-subunit CsGPA1 interacts with the CLV signaling pathway to regulate floral organ number and fruit shape in cucumbers. The study, published in Horticulture Research, highlights the role of CsGPA1 in modulating the expression of key regulatory genes.
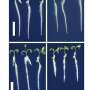
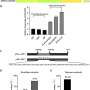
The study employed CRISPR/Cas9 gene editing to create cucumber mutants for CsCLV3, CsCLV1, and CsGPA1. Disrupting CsCLV3 and CsCLV1 led to flowers with additional organs and shorter, broader fruits. The research revealed that CsGPA1 interacts with CsCLV1, and its disruption resulted in similar phenotypes, highlighting its role in the CLV signaling pathway.
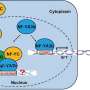
Detailed analysis showed that CsGPA1 influences the expression of WUSCHEL (WUS) and CRABS CLAW (CRC), crucial regulators of SAM size and fruit elongation. Specifically, CsGPA1 represses CsWUS expression and activates CsCRC transcription, thereby controlling floral organ number and promoting fruit elongation.
These findings identify CsGPA1 as a new component in the CLV pathway, offering insights into the genetic regulation of flower and fruit development in cucumbers, and providing potential targets for crop breeding improvements.
Dr. Xiaolan Zhang, one of the study's senior researchers, stated, "Our discovery of CsGPA1's interaction with the CLV signaling pathway provides a novel understanding of how floral and fruit traits are regulated in cucumbers. This could significantly impact future crop breeding programs by allowing more precise manipulation of fruit shape and size."
The identification of CsGPA1 as a key regulator in the CLV signaling pathway opens new avenues for genetic manipulation in crop breeding. By targeting this gene, breeders can potentially develop cucumber varieties with desired floral and fruit traits, improving both yield and quality.
This research not only enhances our understanding of plant development but also provides practical applications for agricultural advancements.
More information: Lijie Han et al, Heterotrimeric Gα-subunit regulates flower and fruit development in CLAVATA signaling pathway in cucumber, Horticulture Research (2024). DOI: 10.1093/hr/uhae110
Journal information: Horticulture Research
Provided by China Agricultural University