This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Hundreds of new genome sequences fill gaps in the fruit fly tree of life

A multitude of new genomic sequence data fills major gaps in the fruit fly tree of life, Bernard Kim from Stanford University, US, and colleagues report in the open-access journal PLOS Biology, publishing July 18.
Fruit flies are classic model organisms in biological research and were among the first species to have their whole genome sequenced. With over 4,400 species, the diversity of the fruit fly family could offer insights into evolutionary patterns and processes. But only a fraction of these species have their genome sequenced, and most published fruit fly genome sequences are from a very limited set of species with representative inbred laboratory strains.
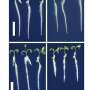
To address this, researchers sequenced the genomes of 179 fly species in the Drosophilidae family, including wild-caught flies, preserved museum specimens and laboratory-reared strains.
Using a hybrid sequencing approach that combines the newest short- and long-read sequencing technologies, they were able to produce low-cost, high-quality genome sequences from limited material.
They used the new genome sequences and previously published data to produce a phylogenetic tree for 360 species in the Drosophilidae family, refining our understanding of the evolutionary relationships of these species. They also aligned nearly 300 fruit fly genomes as an open-source tool for future comparative genomics research, such as a whole-genome alignment.

While large-scale sequencing efforts for larger organisms such as mammals are well underway, this study demonstrates that genome sequencing for small organisms such as individual flies—even those preserved in museums for up to two decades—is now possible.
The authors add, "It is now entirely feasible to think about assembling genomes for hundreds or thousands of species, even on the research budget of a single lab. This kind of large-scale, clade-level sampling will provide us with an unprecedented level of resolution for studying the genome sequences of diverse groups like fruit flies and beyond, which is sure to improve our understanding of the evolutionary process."
More information: Kim BY, Gellert HR, Church SH, Suvorov A, Anderson SS, Barmina O, et al. (2024) Single-fly genome assemblies fill major phylogenomic gaps across the Drosophilidae Tree of Life. PLoS Biology (2024). DOI: 10.1371/journal.pbio.3002697
Journal information: PLoS Biology
Provided by Public Library of Science