This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Using reference genome of the species itself is optimal for SNP calling, finds study

With the advent of next-generation sequencing technologies, restriction-site associated DNA sequencing (RAD-seq) has become a mainstream method for rapidly obtaining high-density single nucleotide polymorphisms (SNPs) in organisms due to its independence from reference genomes. However, few studies have examined the impact of using closely related species as reference genomes versus using the species itself as a reference genome.
In a study published in Plant Science, researchers from the Xishuangbanna Tropical Botanical Garden (XTBG) of the Chinese Academy of Sciences evaluated the benefits and limitations of both reference-based approaches, using a closely related species as the reference genome versus using the species itself as the reference genome.
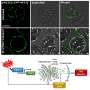
Using the bioinformatics software STACKS, the researchers investigated the effect of using different reference genomes on SNP calling. They utilized RAD-seq data from 242 individuals of Engelhardia roxburghiana, a tropical tree in the walnut family (Juglandaceae). They focused on two different reference genomes: using a closely related species (i.e., Pterocarya stenoptera) as the reference genome, and using the species itself (i.e., Engelhardia roxburghiana) as the reference genome.
They found a significant difference in the number of SNPs obtained between using the species itself as the reference genome and using a closely related species as the reference genome, with the former producing significantly more SNPs than the latter.
"This result indicates that choosing the species itself as the reference genome is the optimal solution for SNP calling," said Li Jie of XTBG.
The researchers suggest that reference genomes of closely related species can be used when the species itself is not available as a reference. It is recommended to avoid using species with a distant phylogenetic relationship.
"Our study contributes to enriching the understanding of the impact of SNP acquisition when using different reference genomes," said Meng Honghu, corresponding author of the study.
More information: Pei-Han Huang et al, Different reference genomes determine different results: Comparing SNP calling in RAD-seq of Engelhardia roxburghiana using different reference genomes, Plant Science (2024). DOI: 10.1016/j.plantsci.2024.112109
Provided by Chinese Academy of Sciences