This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
proofread
The hidden drivers of evolution: Transposable elements in Rosaceae genomes
Transposable elements are mobile DNA sequences that play a crucial role in plant genome architecture and gene regulation. They drive genome size variation and affect gene expression by altering regulatory networks. Despite their significance, the diverse and dynamic roles of transposable elements (TEs) are not well understood.
To address these complexities, in-depth studies are necessary to explore TE activity, distribution, and their effects on gene regulation. This research focuses on the Rosaceae family, aiming to uncover the impact of TEs on genome evolution and functional diversity.
Conducted by a team from Northwest A&F University, this study was published on April 26, 2024, in Horticulture Research. The researchers investigated transposable elements in 14 Rosaceae genomes, examining their distribution, transposition activity, expression patterns, and influence on differentially expressed genes. The goal is to provide comprehensive insights into how TEs shape genome evolution and gene regulation in Rosaceae.
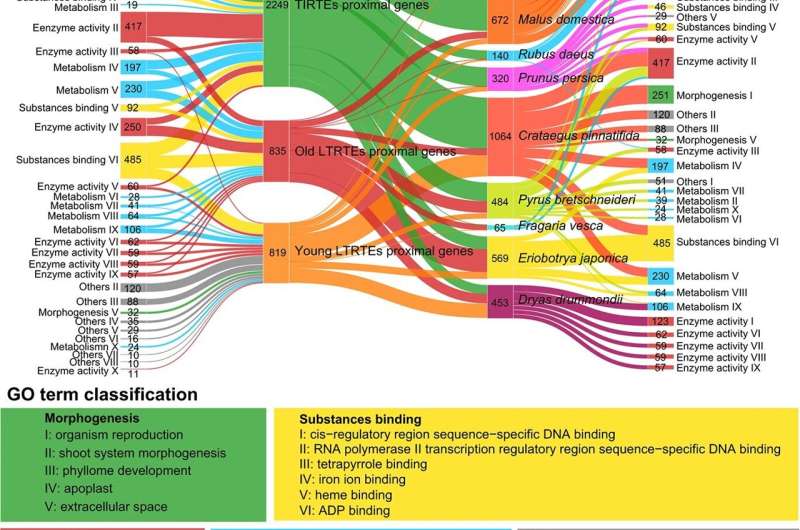
The study analyzed 14 representative Rosaceae genomes, revealing distinct evolutionary patterns of long terminal repeat retrotransposons (LTR-RTs) and their effects on genome size and gene expression. Genes near recent TE insertions were associated with adversity resistance, while those near older insertions were linked to morphogenesis, enzyme activity, and metabolic processes.
The research highlights diverse responses of LTR-RTs to various conditions, demonstrating their significant role in plant genome evolution. Additionally, 3,695 pairs of syntenic differentially expressed genes (DEGs) near TEs in apple varieties were identified, suggesting that TE insertions contribute to varietal trait differences. This analysis provides new insights into the role of TEs in shaping the genomic landscape and regulatory mechanisms within the Rosaceae family.
Dr. Yaqiang Sun, one of the corresponding authors, stated, "Our findings underscore the pivotal role of transposable elements in shaping the genomic architecture and functional diversity within the Rosaceae family. This research provides a valuable resource for future studies on genome evolution and the regulatory mechanisms of TEs."
The insights from this study have significant implications for plant breeding and genetic engineering. Understanding the role of TEs in gene regulation and genome evolution can help develop new strategies for crop improvement, particularly in enhancing stress resistance and optimizing desirable traits in Rosaceae species. This research also sets the stage for further exploration of TEs in other plant families, contributing to a broader understanding of plant genome dynamics.
More information: Ze Yu et al, Transposable elements in Rosaceae: insights into genome evolution, expression dynamics, and syntenic gene regulation, Horticulture Research (2024). DOI: 10.1093/hr/uhae118
Journal information: Horticulture Research
Provided by TranSpread