This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
proofread
Researchers unveil high-quality genome of Rubus rosaefolius
Rubus rosaefolius, a red raspberry, is valued for its nutritional and medicinal properties, containing bioactive compounds like anthocyanins. Despite its potential, there is limited genomic information available.
Previous studies have focused on taxonomy, phytochemistry, and pharmacology. Due to these gaps, an in-depth study of the genomic and molecular basis of important biological properties in Rubus species is necessary.
A study, conducted by researchers from Kaili University and the Botanic Garden of Guizhou Province, published in Horticulture Research, presents a high-quality chromosome-level draft genome of Rubus rosaefolius.
By sequencing and analyzing the genome and transcriptomes, the study explores the evolutionary dynamics and molecular mechanisms underlying anthocyanin biosynthesis in this red raspberry.
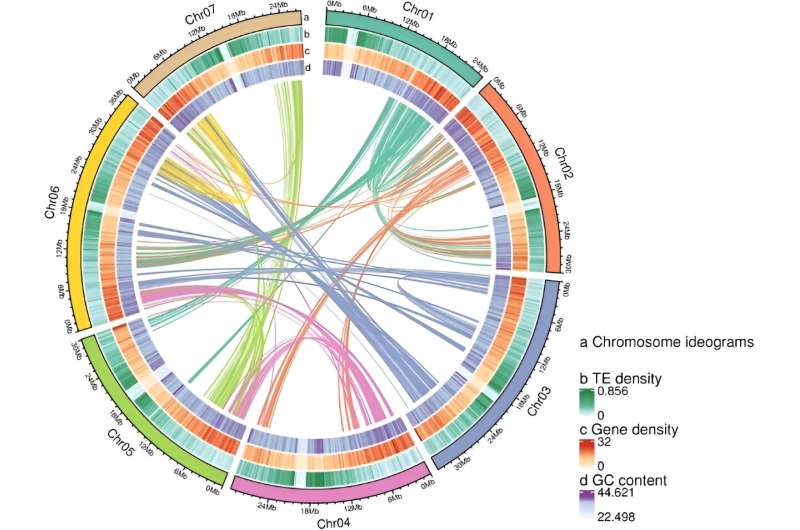
The genome sequencing revealed 131 assembled scaffolds, with 70 anchored to seven pseudo chromosomes, covering 99.33% of the estimated genome size. The study identified a whole-genome duplication event shared among Rosaceae family members, leading to 5090 detectable duplicated gene pairs, with approximately 75% undergoing purifying selection.
Various anthocyanins were detected in the berries, with concentrations increasing significantly during ripening. Key structural genes such as RrDFR, RrF3H, RrANS, and RrBZ1 were identified as crucial in anthocyanin biosynthesis.
The expression of these genes correlated with the accumulation of major anthocyanins like pelargonidin-3-O-glucoside and pelargonidin-3-O-(6′′-O-malonyl) glucoside. Additionally, transcription factors and methylase-encoding genes were found to regulate anthocyanin biosynthesis by targeting structural genes.
Dr. Yunsheng Wang, a lead researcher from Kaili University, said, "Our findings provide comprehensive insights into the genomic evolution and molecular mechanisms of anthocyanin biosynthesis in Rubus rosaefolius. This knowledge is instrumental for the targeted domestication and breeding of Rubus species, enhancing their nutritional and commercial value."
The research paves the way for developing Rubus cultivars with enhanced anthocyanin content, potentially revolutionizing the fruit's role in dietary health and contributing to the advancement of berry breeding programs globally.
More information: Yunsheng Wang et al, Chromosome-scale genome, together with transcriptome and metabolome, provides insights into the evolution and anthocyanin biosynthesis of Rubus rosaefolius Sm. (Rosaceae), Horticulture Research (2024). DOI: 10.1093/hr/uhae064
Journal information: Horticulture Research
Provided by TranSpread