This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Medicinal plant research unveils the genetic blueprint of Chaenomeles speciosa
Chaenomeles speciosa (2n=34), a diploid species within the Rosaceae family, has been traditionally used in Chinese medicine for its various health benefits. To date, the lack of genomic sequence and genetic studies has impeded efforts to improve its medicinal value.
Recent studies have identified C. speciosa fruits as sources of beneficial metabolites like triterpenoids and flavonoids, emphasizing the need for further investigations to understand the phytochemical basis of its medicinal uses. However, while genomic research on other Rosaceae members has progressed, C. speciosa remains unsequenced, limiting our understanding of its genetic makeup and the biosynthesis of its active compounds.
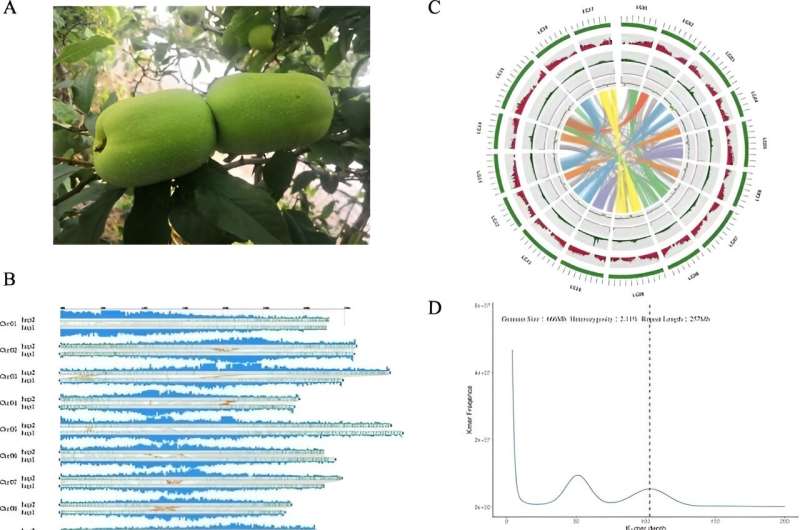
Horticulture Research published research titled "A telomere-to-telomere reference genome provides genetic insight into the pentacyclic triterpenoid biosynthesis in Chaenomeles speciosa." In this study, the genome of C. speciosa was sequenced using a hybrid approach that combined short reads, HiFi, and Hi-C sequencing techniques, revealing a genome size of approximately 650.4 Mb with a heterozygosity rate of 2.1%.
Utilizing HiFi reads, the assembly process resulted in a primary assembly of 167 contigs with a significant N50 length, which was further scaffolded using Hi-C sequencing data to achieve a chromosome-level assembly of 17 pseudo-chromosomes. The analysis also delved into the haplotype-specific assemblies, revealing differences in genome size, contig and scaffold N50 values, and GC content between the two haplotypes identified, Hap 1 and Hap 2. Further, the research uncovered significant genetic variation, including SNPs and InDels, between these haplotypes, indicating a rich genetic diversity within C. speciosa.
Gene family analysis elucidated the presence of species-specific genes and provided insights into the expanded and contracted gene families, highlighting the plant's unique genetic makeup and its implications for secondary metabolite production. The genome's quality was validated through various metrics, including BUSCO analysis, which confirmed its completeness and accuracy.
Additionally, the study offered a comparative perspective with other species, revealing synteny relationships and evolutionary insights, particularly regarding whole-genome duplication events and the expansion of transposable elements, which were instrumental in the genome's evolution.
This assembly facilitates a detailed understanding of the genetic basis for the biosynthesis of oleanolic and ursolic acids, characteristic chemical markers of C. speciosa with medicinal value. The identification and expansion of gene families related to these compounds underscore the plant's potential for producing health-beneficial metabolites.
Overall, this genomic resource sets a foundation for future genetic studies and breeding efforts aimed at enhancing the medicinal properties of C. speciosa, marking a significant advancement in the genetic research of this medicinal and edible plant.
More information: Shaofang He et al, A telomere-to-telomere reference genome provides genetic insight into the pentacyclic triterpenoid biosynthesis in Chaenomeles speciosa, Horticulture Research (2023). DOI: 10.1093/hr/uhad183
Provided by NanJing Agricultural University





















