This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Almond genome study paves way for improved breeding strategies
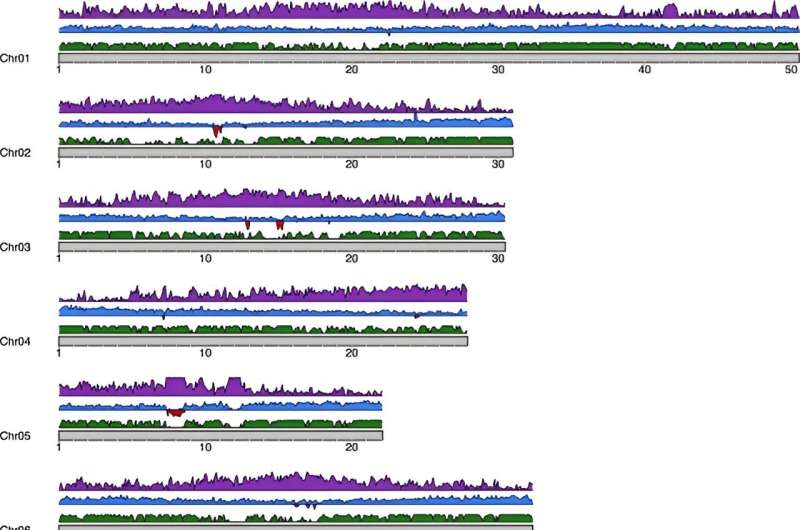
A recent study has unlocked the genetic complexities of almonds, highlighting a significant degree of heterozygosity and the influence of structural variants on gene expression. The research rigorously delineates allele-specific expression patterns, offering vital insights into the genetic traits that govern almond variability. These findings are crucial for refining breeding techniques and bolstering crop resilience.
Almonds exhibit notable genetic diversity, crucial for agricultural advancements. Insights from the phased genome assembly of the "Texas" cultivar illuminate the effects of structural variations on gene expression, broadening our comprehension of the almond's genetic framework. The imperative for comprehensive investigations into these genetic details is apparent, as they are key to harnessing the genetic potential of almonds and enhancing cultivar traits through strategic breeding practices.
Published in Horticulture Research on April 9, 2024, by the Centre for Research in Agricultural Genomics (CRAG), this study delves into the phased genome of the "Texas" almond cultivar. It scrutinizes how high heterozygosity and structural variants modulate allele-specific expression. These revelations deepen our genetic understanding of almonds and lay the groundwork for future agricultural and breeding breakthroughs.
Leveraging advanced genomic sequencing techniques, researchers combined 260X PacBio long reads with 172X Hi-C Illumina short reads to construct a detailed phased genome of the "Texas" almond. This methodology exposed the significant role of heterozygous transposable element insertions in regulating gene expression. The enhanced genome assembly, with a 13% increase in sequence data over previous versions, provides intricate insights into genetic interactions and functions.
The study underscores the critical role of structural variants in trait variability, offering invaluable perspectives that could inform targeted almond breeding strategies to improve desirable traits.
Dr. Josep M. Casacuberta, a leading researcher in the study, emphasized, "This phased genome assembly represents a major leap in our comprehension of the almond genome. It not only clarifies the genetic complexity but also paves the way for new biotechnological applications in almond breeding and agriculture."
The findings from this phased genome assembly carry significant implications for almond breeding programs, introducing innovative strategies to enhance traits such as disease resistance and yield. Furthermore, the study's methods could be adapted to other crop species, potentially transforming agricultural genomics.
By elucidating the specific interactions between structural variants and gene expression, researchers can more effectively devise interventions to boost crop resilience and productivity.
More information: Raúl Castanera et al, A phased genome of the highly heterozygous 'Texas' almond uncovers patterns of allele-specific expression linked to heterozygous structural variants, Horticulture Research (2024). DOI: 10.1093/hr/uhae106
Journal information: Horticulture Research
Provided by NanJing Agricultural University