This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Integrating non-additive GWAS with historical dissemination to illuminate nut traits and blooming time in almonds
Modern breeding focuses on genetic analyses and germplasm management and dominates in altering crop genomes, but often neglects non-additive genetic effects that are essential for understanding traits. Almond [Prunus dulcis Miller (D.A. Webb)] has significant economic and genetic research value, but current studies face limitations, including a narrow focus on additive models and limited germplasm diversity.
Expanding research to include a wider range of germplasms and non-additive effects could uncover deeper insights into almond domestication and genetic diversity, revealing important alleles and QTLs developed over thousands of years.
Horticulture Research published research titled "Almond population genomics and non-additive GWAS reveal new insights into almond dissemination history and candidate genes for nut traits and blooming time."
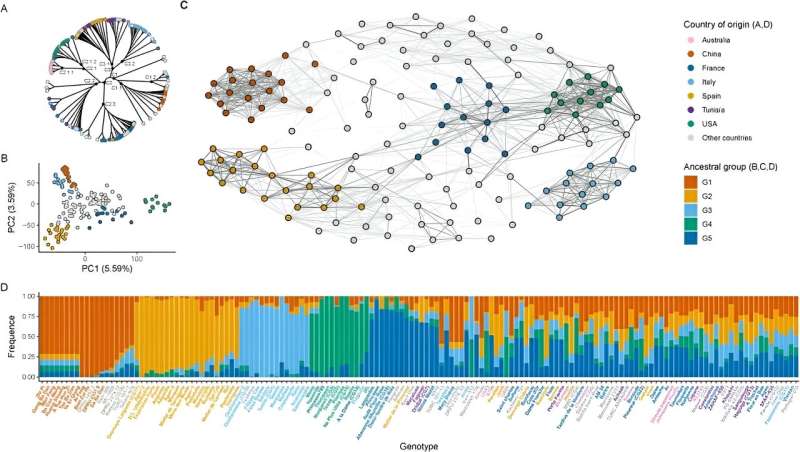
This study utilized a comprehensive approach combining population structure analysis, additive kinship, phylogenetic tree construction, and Principal Compound Analysis (PCA) to elucidate the genetic structure as well as additive and non-additive associations between genotypes and phenotypes of 243 almond accessions. Genetic structure analysis divided these varieties into five distinct ancestral groups, all of which were composed of varieties with a common origin.
One of these groups was composed exclusively of Spanish accession, whereas the others consisted mainly of varieties from China, Italy, France, and the United States. PCA underscored the separation of ancestral groups and highlighted the central positioning of admixed accessions. These results are consistent with archaeological and historical evidence that the spread of modern almonds occurred in four phases: Asia, the Mediterranean, Californian, and the Southern Hemisphere.
Genomic analysis revealed differences in allele frequencies related to domestication and selection pressures, pinpointing regions of interest on chromosomes 1, 4, and 5. Notably, linkage disequilibrium decay was relatively short, emphasizing the precision in locating candidate genes for key almond traits.
The study identified 13 independent QTLs for nut weight, crack-out percentage, double kernel percentage, and blooming time, which showed mainly non-additive effects. Through candidate gene analysis, this research proposed Prudul26A013473 as a candidate gene for the main QTL of crack-out percentage, Prudul26A012082 and Prudul26A017782 for the QTL of double kernel percentage, and Prudul26A000954 for the QTL of blooming time. This is significant for understanding genotype-phenotype interactions in almonds and potentially other Prunus species.
In conclusion, this study combines genetic analyses with historical and archaeological evidence to provide a comprehensive view of almond genetic diversity and reveal the complex history of its worldwide dissemination. The identification of QTLs and candidate genes offers valuable insights for breeding and conservation strategies, emphasizing the importance of considering non-additive genetic effects in crop improvement programs.
More information: Felipe Pérez de los Cobos et al, Almond population genomics and non-additive GWAS reveal new insights into almond dissemination history and candidate genes for nut traits and blooming time, Horticulture Research (2023). DOI: 10.1093/hr/uhad193
Provided by NanJing Agricultural University