This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Telomere-to-telomere genome assembly of melon provides a high-quality reference for meta-QTL analysis
Melon, with its significant economic value and extensive phenotypic diversity, has been cultivated globally for over 4,000 years, featuring two main subspecies that have undergone independent domestication processes. Recent advancements have led to the assembly of several high-quality melon genomes, enhancing our understanding of genetic diversity and improving genetic mapping.
However, the complexity of the melon genome and the limitations of sequencing technologies have left many areas fragmented and incomplete, hindering a full comprehension understanding of its genome structure and related biological function. Additionally, while quantitative trait loci (QTL) mapping has advanced, identifying candidate genes remains challenging due to variable environments, genetic backgrounds, and large mapping intervals.
Horticulture Research published research titled "Telomere-to-telomere genome assembly of melon (Cucumis melo L. var. inodorus) provides a high-quality reference for meta-QTL analysis of important traits."
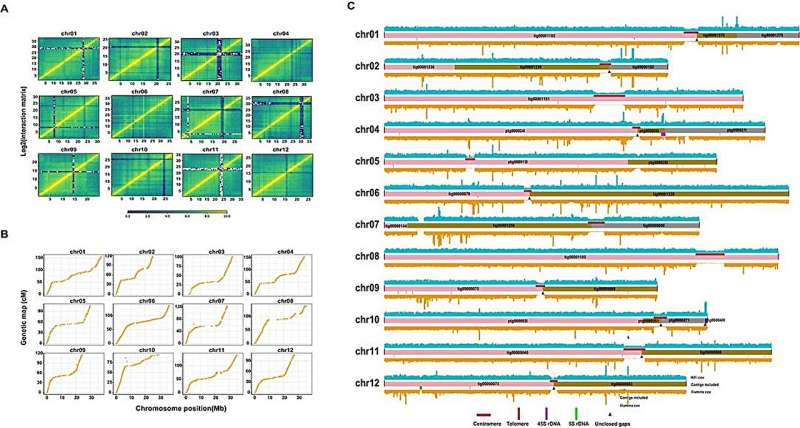
In this study, the elite melon landrace Kuizilikjiz (Cucumis melo L. var. inodorus) was selected for de novo telomere-to-telomere (T2T) genome assembly, incorporating a comprehensive projection of previously published QTLs. Utilizing a combination of short reads, PacBio high-fidelity long reads, Hi-C data, and a high-density genetic map, a high-quality genome of 379.2 Mb with an N50 of 31.7 Mb was assembled, including the identification of centromeres and telomeres on each chromosome and revealing a significant number of structural variations compared to other melon genomes.
Specifically, the assembly process involved initial Illumina sequencing for genomic characteristics, followed by PacBio sequencing for contig assembly, Hi-C sequencing for scaffolding, and genetic linkage mapping for validation and gap closure, resulting in a highly contiguous and complete assembly with low base-level error.
Furthermore, a total of 1,294 QTLs from 67 published studies were collected and projected onto this genome, leading to the identification of 20 stable meta-QTLs, significantly narrowing down the mapping intervals of initial QTLs and aiding in candidate gene identification.
This integration of QTLs onto a high-quality T2T genome assembly not only enhances the resources available for melon genetic research but also provides valuable information for gene cloning related to important traits.
Overall, the comparative genomic analysis underscored the uniqueness of the Kuizilikjiz genome within melon genetic resources, and the meta-QTL analysis demonstrated its efficacy in refining candidate gene predictions, marking a significant advancement in melon genomic research and breeding efforts.
More information: Minghua Wei et al, Telomere-to-telomere genome assembly of melon (Cucumis melo L. var. inodorus) provides a high-quality reference for meta-QTL analysis of important traits, Horticulture Research (2023). DOI: 10.1093/hr/uhad189
Journal information: Horticulture Research
Provided by NanJing Agricultural University