This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Research team develops universal and accurate method to calculate how proteins interact with drugs
A research team from the Institute of Organic Chemistry and Biochemistry of the Czech Academy of Sciences / IOCB Prague has developed a novel computational method that can accurately describe how proteins interact with molecules of potential drugs and can do so in a mere tens of minutes. This new quantum-mechanical scoring function can thus markedly expedite the search for new drugs. The research has been published in the journal Nature Communications.
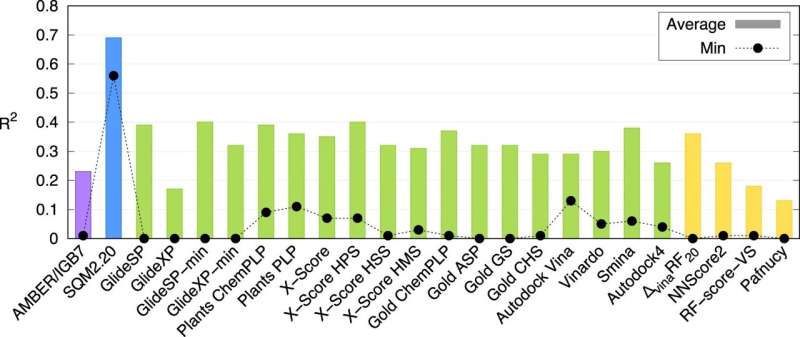
The study demonstrates that this is the first universally applicable method of its kind. IOCB Prague computational experts tested it on 10 proteins of different levels of structural complexity, each binding a large variety of small molecules (usually referred to as ligands). They then compared their results not only with those of other corresponding methods, but also with findings of laboratory experiments, and both comparisons turned out very favorably.
"Of course, we are not the only ones working on this. There are several such methods. Usually, however, their speed is offset by low accuracy whereas more accurate calculations can take several days. Our methods are unique in that they can process information about large molecular systems within tens of minutes while retaining the benefits of much more demanding quantum-mechanical calculations," explains Jan Řezáč, corresponding author of the article from the Non-Covalent Interactions group led by Prof. Pavel Hobza.
Experts from this group have been studying intermolecular interactions for a long time. In this research they focus mainly on biomolecules, and the results of their work directly bear on the computer-aided design of drugs. The reason is that when scientists work toward a new drug, they often look for molecules that bind strongly to a particular protein.
Identifying them, however, is akin to finding needles in a haystack, as large numbers of molecules have to be tested to set apart those that show promise. This considerably slows down the discovery of medicinal substances and makes it more expensive. By predicting the strength of protein–ligand binding, and thus singling out molecules that best satisfy a defined set of criteria, computational chemists spare the work of experimenters, which, in turn, significantly accelerates drug discovery.
More information: Adam Pecina et al, SQM2.20: Semiempirical quantum-mechanical scoring function yields DFT-quality protein–ligand binding affinity predictions in minutes, Nature Communications (2024). DOI: 10.1038/s41467-024-45431-8
Journal information: Nature Communications