Computer simulation of receptors reveals a new ligand-binding site
Using a computer simulation of an important receptor, EPFL scientists have discovered a novel binding site for natural ligands and drugs. The new site might be present on other receptors and can be exploited in novel treatments for multiple diseases.
Most biological processes in a cell go through receptors. These are specialized proteins that are activated when a ligand binds to them. Ligands can be all sorts of molecules (e.g. hormones, nucleic acids, neurotransmitters etc), and by binding receptors – and other proteins – they run complex processes such as cell maintenance, immune responses, genetics and others.
These normal cell processes involve complex "dominoes" of biochemical signals that are carried across the cell through protein-ligand interactions. On the other hand, they also lie at the core of a vast number of diseases. An entire branch of research known as "receptor pharmacology" is dedicated to exploiting these interactions with synthetic ligands (drugs), attempting to find ligand-binding sites that can exploited as a drug target.
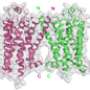
Now, scientists led by Professor Horst Vogel at EPFL have developed a computer simulation of the widespread muscarinic acetylcholine receptor. Specifically, the scientists examined the M3 and M4 subtypes of the receptor, which are majorly involved in the function of the lungs (M3) and the central nervous system (M3 and M4).
The receptors belong to the large family of so-called "G protein-coupled receptors" (GPCRs), which generally detect signals coming from outside the cells, such as light, hormones, or neurotransmitters. Upon activation, GPCRs change their structure in such a way that they can bind and activate other proteins inside the cell and ultimately turn on the appropriate process.
GPCRs are the target of more than a third of modern therapeutic compounds, meaning that finding new ligand binding sites can help design more efficient GPCR-targeted drugs.
The team used what are known as "molecular dynamics simulations", which is a method for simulating studying the physical movements of atoms and molecules in a computer. The method can reveal details down to the level of individual atoms, thus offering a high-resolution way to look at how various ligands bind on the receptor – and, more importantly, where.
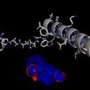
The study revealed a new binding site on the acetylcholine receptors, which can be exploited pharmacologically to understand ligand binding and activation processes. When a ligand binds to the new site, it causes the entire site to expand. The site seems capable of binding small ligands and cause different effects to the receptor than the "main" ligand would.
Looking at more than 200 ligand-bound GPCR structures, the researchers discovered that most ligands bind the traditional ("orthosteric") sites on the receptors. However, one receptor that binds leukotriene (LTB4) and directs immune cells to sites of infection seemed to bind a "double" ligand, which binds the new site discovered in the two acetylcholine receptors. The new site was detected in many others of the over 200 receptors that the scientists examined.
The study shows that the new binding site might exist in other GPCRs, opening a new opportunity for GPCR drug discovery. "The study shows the power of computational methods to resolve in atomic detail central receptor-mediated signaling reactions," says Horst Vogel. "The challenging next step is to use computational methods to design novel compounds that would fit into the newly found binding sites to activate or deactivate the receptor in a defined mode and thus design novel medicines."
More information: H. C. Stephen Chan et al. Exploring a new ligand binding site of G protein-coupled receptors, Chemical Science (2018). DOI: 10.1039/C8SC01680A
Journal information: Chemical Science
Provided by Ecole Polytechnique Federale de Lausanne