This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
New findings on Bacteroides thetaiotaomicron physiology under bile stress
Researchers from the Würzburg Helmholtz Institute for RNA-based Infection Research (HIRI) deployed CRISPR interference for the first time for the functional characterization of the gut mutualist Bacteroides thetaiotaomicron. They identified a small ribonucleic acid (sRNA) that modulates the microorganisms' growth in the presence of bile.
The findings should contribute to a better understanding of the bacterium's lifestyle in its native environment, the human intestine, and to the development of clinical applications. The study was published in the Proceedings of the National Academy of Sciences.
Bacteroides thetaiotaomicron is an abundant member of the human intestinal microbiota. The bacterium supports the digestion of polysaccharides and is of paramount importance for human health, but it can also cause or promote infections. As a model organism, B. thetaiotaomicron is increasingly being researched, however, its gene functions are still poorly understood. The latter is particularly true for the noncoding genes which are transcribed into small, noncoding ribonucleic acids (sRNAs for short) without being translated into proteins.
"Our gut mutualists are affecting our health as well as diseases, but there is barely any knowledge as to the function of noncoding genes," says Alexander Westermann.
Given the important functions that the noncoding DNA regions have in infection by pathogens, it can be assumed that they play a similar key role in beneficial bacteria such as Bacteroides, says Westermann.
Westermann initiated the study and is a research group leader at the Würzburg Helmholtz Institute for RNA-based Infection Research (HIRI), a site of the Braunschweig Helmholtz Center for Infection Research (HZI) in cooperation with the Julius-Maximilians-Universität Würzburg.
A lack of specific therapies
"Microbiota-centric interventions are limited by our incomplete understanding of the gene functions of many of its constituent species. While the importance of sRNA genes in bacteria has been recognized, tools for their global functional characterization have been lacking," says Westermann.
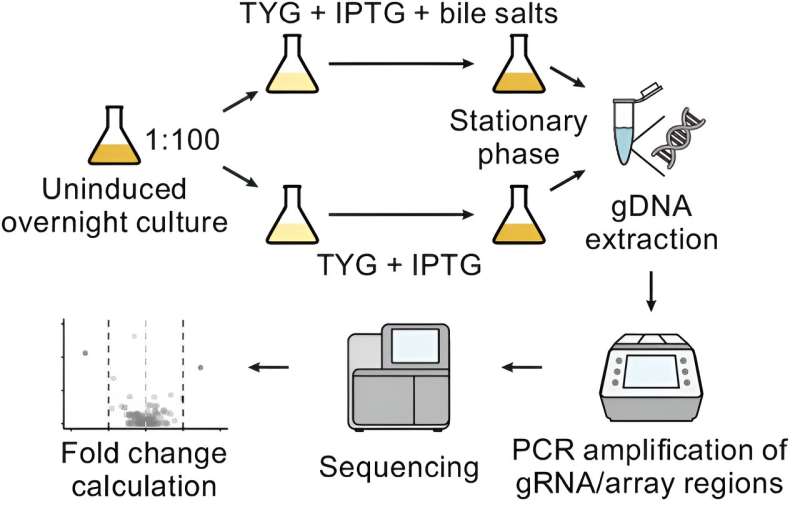
In collaboration with Chase Beisel's department at HIRI, the scientists from the Westermann lab have now deployed a molecular biochemical tool known as CRISPR interference (CRISPRi for short) to tackle this problem. Using specifically engineered guide RNAs, CRISPRi blocks the expression of selected genes, virtually knocking them out.
"Despite ongoing attempts, to our knowledge CRISPRi has not previously been used for a systematic functional screening of Bacteroides thetaiotaomicron genes," says Gianluca Prezza, first author of the study and Ph.D. student in Westermann's lab.
A previously uncharacterized ribonucleic acid
The researchers employed CRISPRi to generate a targeted knockdown library of the intergenic sRNA repertoire of these important gut bacteria. In the subsequent screening, they identified a previously uncharacterized sRNA, which regulates genes involved in Bacteroides cell surface assembly and confers enhanced susceptibility to bile salts. Prezza states, "Suppressing the identified sRNA, called BatR, increases Bacteroides resilience to bile stress."
Overall, the guide RNA library presented bears potential to systematically uncover the gene functions of Bacteroides under a variety of experimental conditions. "Our work illuminates the benefits of CRISPRi for the functional characterization of sRNAs and lays the ground for a targeted gene knockdown in these abundant human microbiota members," says Westermann. At the same time, the study provides the foundation for the replication of this approach in other bacterial species.
More information: Gianluca Prezza et al, CRISPR-based screening of small RNA modulators of bile susceptibility in Bacteroides thetaiotaomicron, Proceedings of the National Academy of Sciences (2024). DOI: 10.1073/pnas.2311323121. www.pnas.org/doi/10.1073/pnas.2311323121
Journal information: Proceedings of the National Academy of Sciences
Provided by Helmholtz Association of German Research Centres