This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Research quantifies how millions of cells in zebrafish embryos are affected by key gene alterations
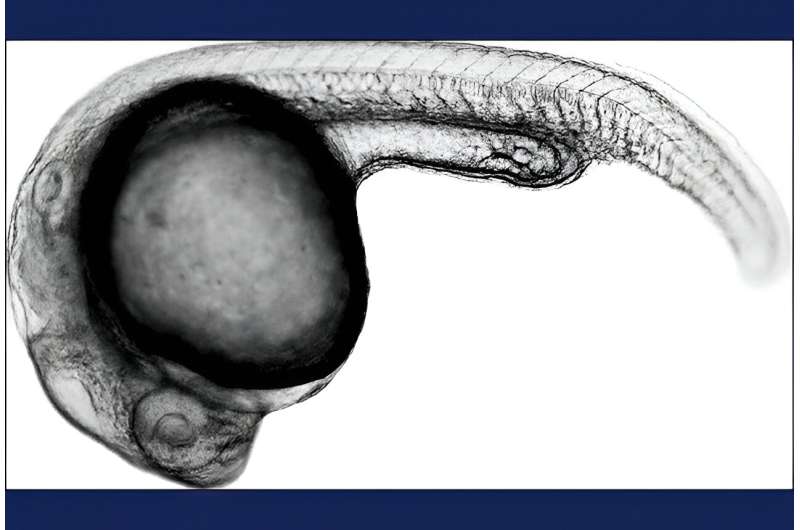
Seattle researchers have developed a technique to quantify the changes in gene activity that occur throughout zebrafish embryos in response to specific edits to key genes. The approach makes it possible to quantify gene activity and the effect of genetic variations in millions of cells in thousands of embryos during their development.
The advance will speed the study of normal embryonic development and advance the understanding of how mutations in specific genes affect cells throughout an embryo and cause disease, the researchers said.
"Now we can use the zebrafish to determine how the loss of a particular gene affect all the cells in an organism," said co-lead author Lauren Saunders, of EMBL Heidelberg, Germany. She was recently a postdoctoral researcher in the laboratory of Cole Trapnell, who was the study's senior co-author. Trapnell is an associate professor of genome sciences at the University of Washington School of Medicine. Sanjay R. Srivastan, now a Fred Hutch Cancer Center investigator, and David Kimelman, an emeritus professor biochemistry at the UW medical school, were co-lead and co-senior authors, respectively.
"This study provides important clues about what the gene is doing and where, and will perhaps someday show how different therapies could prevent or treat related genetic disorders," Saunders said.
The study results were published on Nov. 15 in the journal Nature in the paper, "Embryo-scale reverse genetics at single-cell resolution."
Previous research had mapped gene-expression differences in cells of zebrafish embryos, but these maps did not reveal the differences in gene expression that occur between individual embryos. The earlier maps also lacked tightly spaced timepoints during the period of mid-late-stage embryogenesis. In addition, past results represented the gene-expression profile of a wild-type version of the organism, or a couple of genetic perturbations, only at a single timepoint.
In the new study, the researchers labeled the transcriptomes of more than 1,800 embryos. They then tracked the changes in each cell type over time by sampling embryos at 19 points during their development. They also introduced 23 distinct genetic perturbations, which made it possible to see how each mutation affected gene expression and cellular development across all cell types of the organism over time.
To keep track of which cells came from which embryos, the researchers used a technique called sci-Plex, which was developed in the laboratories of Trapnell and Jay Shendure, a UW School of Medicine professor of genome sciences, scientific director of the Brotman Baty Institute for Precision Medicine, and a Howard Hughes Medical Institute Investigator, who also co-authored this study.
With this technique, the researchers first labeled the nuclei of each embryo with short DNA molecules, with a unique sequence per embryo. These sequences served as a barcode that allowed the researchers to identify which cell came from which embryo. With this approach, they tracked more than 3 million cells they analyzed.
To identify which genes were active in each cell type, they took advantage of the fact that when a gene is active, the genetic instructions encoded in its DNA must first be copied into a related molecule called messenger RNA, or mRNA. The cell then uses the mRNA-encoded instructions as a blueprint to synthesize the protein for which the gene codes. When a gene is active, its mRNA levels rise within a cell; when it is inactive, its mRNA level are low or nonexistent. Thus, mRNA levels tell you when a gene is "on" or "off."
Because each cell type has a different function and a different gene-expression signature, it was possible to identify which cells represented different cell types by measuring mRNA alone. As a result, the researchers could quantify not only how gene expression changes in response to genetic perturbations, but also whether certain populations of cells increased or decreased in the embryo.
Saunders said the findings will expand knowledge on normal and abnormal embryonic development, as well as advance the understanding of animal and human evolution.
The researchers found some surprising relationships between the embryonic origin of cells, their final outcome and their gene-expression profiles. By studying genes important for the development of the notochord, a rodlike structure that runs the length of the embryo, they discovered that cells with notochord-like gene expression profiles were instead early skull base cartilage.
"In zebrafish, and in humans, the skull has two embryonic origins," Saunders said. "We found that the cells that become the base of the skull resemble those of the notochord, rather than the rest of the early skull cartilage which includes the face. It is still unclear how the former population evolved, and our data now enables us to make new hypotheses about how the cartilage-producing cells of the skull arose during vertebrate evolution."
More information: Lauren M. Saunders et al, Embryo-scale reverse genetics at single-cell resolution, Nature (2023). DOI: 10.1038/s41586-023-06720-2
Journal information: Nature
Provided by University of Washington School of Medicine