This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Towards a better understanding of early human embryonic development
The onset of embryo-specific gene transcription, also known as embryonic genome activation (EGA), is a crucial step in the developmental journey of an organism. Although EGA has been studied to some extent in mice, human EGA remains largely unexplored, mainly due to the lack of novel in vitro cell models and ethical restrictions on the usage of human embryos.
Thus, cell models resembling the human blastomere stage—when the embryo undergoes a cell duplication process—are necessary to study the earliest stages of human EGA and understand the events that occur during early embryonic development.
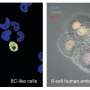
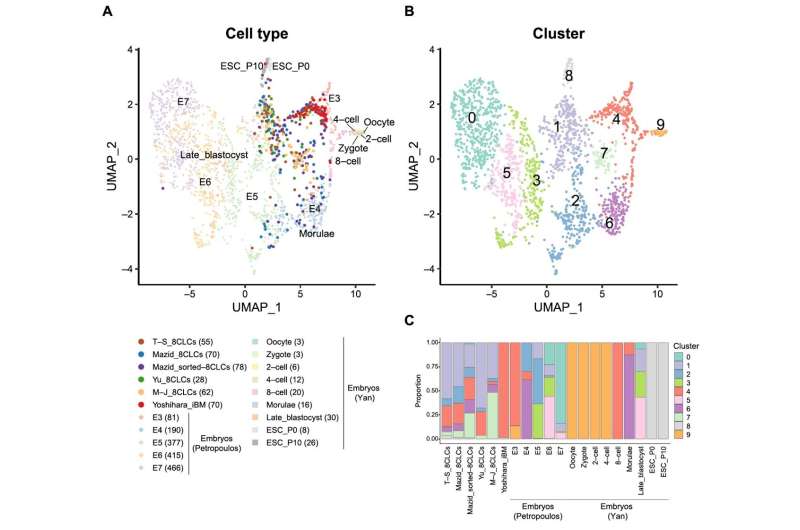
To enable such studies, five independent research groups recently developed different methods to produce human 8-cell-like cells (8CLCs)—a small subpopulation of cells derived from human pluripotent stem cells (hPSCs)—closely resembling the 8-cell-stage embryo. Taubenschmid–Stowers et al. and Moya–Jódar et al. found 8CLCs from naïve hPSCs under two different but specific culture conditions, while Mazid et al. optimized culture conditions to determine the existence of 8CLCs in naïve hPSCs.
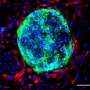
Elsewhere, Yu et al. employed chemical screening to promote the conversion of pre-implantation epiblast-like hPSCs to 8CLCs. Yoshihara et al. reprogrammed induced blastomere-like (iBM) cells from human embryonic stem cells (hESCs) by transient expression of DUX4, a transcription factor activated just after fertilization. Although all research groups identified these cells as 8CLCs using single-cell RNA sequencing (scRNA-seq), the extent of similarities or differences among these 8CLC populations remains unknown.
In a new study, Associate Professor Masahito Yoshihara from the Institute for Advanced Academic Research and Graduate School of Medicine at Chiba University, along with Professor Juha Kere from the Department of Biosciences and Nutrition at Karolinska Institutet, Sweden, and University of Helsinki, Finland, set out to bridge this knowledge gap.
They compared the transcriptomic profiles of the 8CLCs reported by the five research groups, including theirs, with each other and with 8-cell-stage blastomeres. Their findings were published in Stem Cell Reports.
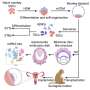
For this comparison, the researchers first integrated the scRNA-seq data of the five developed 8CLCs with two datasets of human pre-implantation embryos—Petropoulos et al. and the Yan et al. datasets. The Yan et al. dataset included primed hESCs and embryos, while the Petropoulos et al. dataset included data from embryonic day three (8-cell stage) till day seven.
Statistical analysis of the successfully integrated data revealed that the iBM cells reprogrammed by Dr. Yoshihara and his team showed the highest similarity to the 8-cell-stage embryo across both datasets, while the other 8CLCs were heterogenous. These findings were reinforced by the cell type annotation of the 8CLCs using the scRNA-seq data of human pre-implantation embryos as references.
Gene expression analysis of all 8CLCs revealed that EGA genes were highly expressed, with pluripotency gene expression being minimal in iBM cells. The other 8CLCs displayed higher expression of pluripotency genes. The researchers also found compelling evidence suggesting that the origin of 8CLCs, as well as their mode of reprogramming, might affect the final cell properties.
They anticipate that the present findings will trigger more extensive research in the early human embryonic development. As Dr. Yoshihara explains, "The developed cell models will enable us to study the earliest stages of human life without ethical concerns. In addition, reprogrammed cells overcome the constraint of limited study specimens, since they can be produced in large numbers at once."
With further clarity on the mechanism of normal early human development, it may be possible to find new methods for understanding the causes of infertility and improving the success of in vitro fertilization.
More information: Masahito Yoshihara et al, Transcriptomic differences between human 8-cell-like cells reprogrammed with different methods, Stem Cell Reports (2023). DOI: 10.1016/j.stemcr.2023.06.009
Journal information: Stem Cell Reports
Provided by Chiba University