This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
proofread
Research team developing a nano-sized force sensor and improving high-precision microscopy technology
In many cases, cells are very active in their movement and serve as power generators. The ability of cells to produce physical forces is one of the basic functions of the body. When running, for example, the forces generated in the cells cause the muscles to contract and the breath to work. It has been possible to measure even the forces experienced by individual proteins by force sensors developed in the past, but previously intracellular forces and mechanical strains could not have been measured.
Together with the scientists from The Ohio State University OSU, cell biology researchers at Tampere University have developed a force sensor that can be attached to the side of a mechanically responding protein, allowing it to sense forces and strain on the protein within the cell.
The development of the micro-sized sensor began on a conference travel in December 2019.
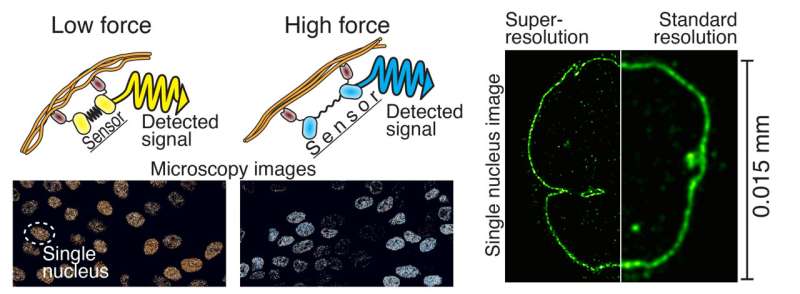
"The power-sensing part is like a rubber band that changes color when stretched. This part is attached to the antibodies at both ends of the rubber band, which bind to the cellular target protein under study. The force or elongation of the studied protein can then be detected under a microscope by following the elongation of the rubber band, i.e. the color it produces," says Teemu Ihalainen, a Senior Research Fellow from BioMediTech at Tampere University.
According to Ihalainen, the force sensor, which is only about twenty nanometers in size, can be easily generalized to a wide range of cell biology research and various target proteins. With the help of the protein biosensor, forces can be measured, for example, in the nuclear membrane, between different proteins, or generally in the cytoskeleton of the cell. It allows the mechanics of the cell to be transformed into visible form for the first time.
There has already been great interest in this technology in various laboratories in Japan, India, Norway and the United States.
Internal forces of the cell provide information on the mechanics of cancer
Cells are subject to forces all the time, both in normal bodily functions and diseases.
As a cancer cell grows and moves, for example, the cells are subjected to mechanical forces. As the cancer spreads e.g., when it enters blood or lymphatic vessels, the cancer cell has to squeeze through narrow gaps in its microenvironment. Thus, cancer cells are subjected to powerful compressive and stretching forces that can break down some of the cells. Damage to the nucleus can alter its genome structure, which in some situations can even be beneficial for the development of cancer.
"With the help of sensors, the mechanics of cancer and related processes can be monitored from a completely new perspective," Ihalainen mentions.
The study was published in Nature Communications.
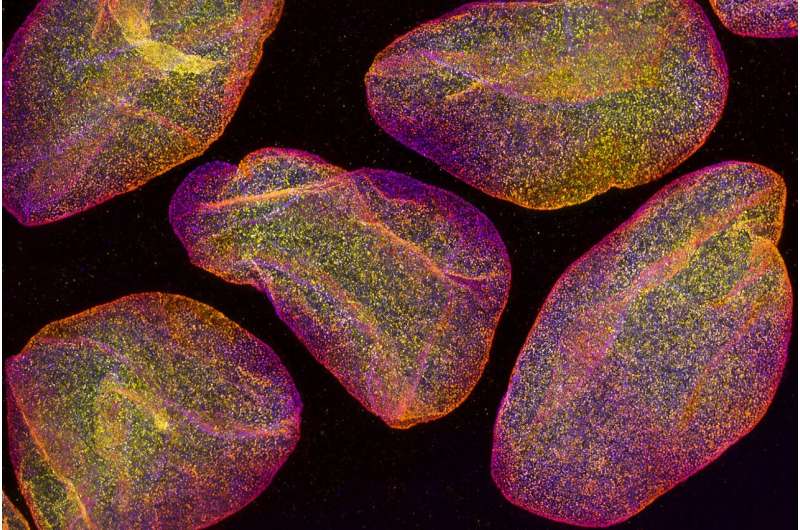
Even the smallest details can be seen with super-resolution microscopy
Another recent study refined expansion microscopy by combining cell biology and signal processing expertise. In addition to cell biology researchers, imaging specialists from the Faculty of Engineering and Natural Sciences at Tampere University and virologists from the University of Jyväskylä participated in the study.
The resolution of light microscopy is limited since the details of small structures in the sample are blurred due to lens-light interactions. However, different techniques of super-resolution microscopy allow for the separation of very small details. One of these techniques is so-called expansion microscopy, the principle of which is to physically enlarge a subject, e.g. a cell, and thus look at the tiny things inside it. In practice, the sample is cast in a soft gel, which can be expanded fourfold or more, and it also enlarges all the details of the sample.
"However, the problem has been that the smaller the details of the cell are examined, the fewer molecules are visible. This means that less signal i.e., information, have been acquired from the sample, and usually there is a lot of noise, a bit like snow on a TV screen," Ihalainen says.
The research group found that the solution to the problem could be repeated fluorescent labeling of the cells. They produced the idea of labeling the target proteins many times to make them appear brighter and provide more information.
"In practice, what we did was to pump more fluorescent molecules to the target proteins as if we were adding reflectors. The simple and easy method greatly improved the resolution and contrast of the image. Noise was also computationally removed from the images, which further increased the image sharpness," he mentions.
Unlike many super-resolution microscopy techniques, expansion microscopy does not require expensive instruments and is easy to implement. The technique developed by the researchers is particularly useful for studying really small details. Looking at the structure of the 120-nanometer Herpes virus, for example, is now possible even with a light microscope. With traditional light microscopy, viruses are visible only as single dots.
The study "Iterative immunostaining combined with expansion microscopy and image processing reveals nanoscopic network organization of nuclear lamina" was published in Molecular Biology of the Cell journal.
More information: Brooke E. Danielsson et al, Nuclear lamina strain states revealed by intermolecular force biosensor, Nature Communications (2023). DOI: 10.1038/s41467-023-39563-6
Elina Mäntylä et al, Iterative immunostaining combined with expansion microscopy and image processing reveals nanoscopic network organization of nuclear lamina, Molecular Biology of the Cell (2023). DOI: 10.1091/mbc.E22-09-0448
Journal information: Nature Communications , Molecular Biology of the Cell
Provided by Tampere University