This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Large-scale genetic modification method reveals the role and properties of duplicated genes in plants
For the first time, researchers from Tel Aviv University have developed a genome-scale technology that makes it possible to reveal the role of genes and traits in plants previously hidden by functional redundancy.
The researchers point out that since the agricultural revolution, man has improved plant varieties for agricultural purposes by creating genetic diversity. But until this recent development, it was only possible to examine the functions of single genes, which make up only 20% of the genome. For the remaining 80% of the genome, made up of genes grouped in families, there was no effective way, on the large scale of the whole genome, to determine their role in the plant.
As a result of this unique development, the team of researchers managed to isolate and identify dozens of new features that had been overlooked until now. The development is expected to revolutionize the way agricultural crops are improved as it can be applied to most crops and agricultural traits, such as increased yield and resistance to drought or pests.
The research was performed by postdoctoral student Dr. Yangjie Hu under the guidance of Prof. Eilon Shani and Prof. Itay Mayrose from the School of Plant Sciences and Food Security at Tel Aviv University's Wise Faculty of Life Sciences. Scientists from France, Denmark, and Switzerland also participated in the research, which was published in Nature Plants.

As part of the research, the team of researchers used the innovative technology "CRISPR" for gene editing and methods from the field of bioinformatics and molecular genetics to develop a new method for locating genes responsible for specific traits in plants.
Prof. Shani says, "For thousands of years, since the agricultural revolution, man has been improving different plant varieties for agriculture by promoting genetic variation. But until a few years ago it was not possible to genetically intervene in a targeted manner, but only to identify and promote desirable traits that were created randomly. The development of gene editing technologies now allows precise changes to be made in a large number of plants."
The researchers explain that despite the development of genetic editing technologies, such as CRISPR, several challenges remained that limited its application to agriculture. One of them was the need to identify as precisely as possible which genes in the plant's genome are responsible for a specific desired trait to cultivate. The accepted method to address this challenge is to produce mutations, that is, to modify genes in different ways and then to examine changes in the plant's traits as a result of the mutation in the DNA and to learn from this about the function of the gene.
Thus, for example, if a plant with sweeter fruit develops, it can be concluded that the altered gene determines the sweetness of the fruit. This strategy has been used for decades with success, but it has a fundamental problem: An average plant such as tomato or rice has about 30,000 genes, and about 80% of them do not work alone but are grouped in families of similar genes.
Therefore, if a single gene from a certain gene family is mutated, there is a high probability that another gene from the same family (actually a copy very similar to the mutated gene) will mask the phenotypes in place of the mutated gene. Due to this phenomenon, called genetic redundancy, it is difficult to create a change in the plant itself and to determine the function of the gene and its link to a specific trait.
The current study sought to find a solution to the problem of genetic redundancy by using an innovative gene editing method called CRISPR. Prof. Mayrose says, "The CRISPR method is based on an enzyme called Cas9 found naturally in bacteria, whose role is to cut foreign DNA sequences. So the enzyme can associate an sgRNA sequence, which identifies the DNA sequence that the enzyme needs to cut. This genetic editing method allows us to design different sgRNA sequences to allow Cas9 to cut almost any gene that we want to change. We wanted to apply this technique to improve the control of creating mutations in plants for the purposes of agricultural improvement, and specifically to overcome the common limitation posed by genetic redundancy."
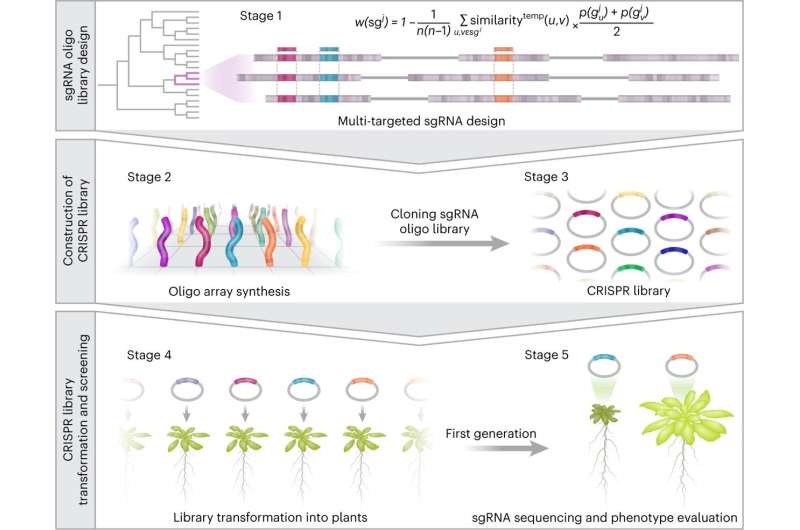
In the first stage, a bioinformatics study was carried out on a computer, which, unlike most studies in the field, initially covered the entire genome. The researchers chose to focus on the Arabidopsis plant, which is used as a model in many studies and has about 30,000 genes. First, they identified and isolated about 8,000 individual genes, which have no family members, and therefore no copies in the genome. The remaining 22,000 genes were divided into families, and for each family appropriate sgRNA sequences were computationally designed.
Each sgRNA sequence was designed to guide the Cas9 cutting enzyme to a specific genetic sequence that characterizes the entire family, with the aim of creating mutations in all family members so that these genes can no longer overlap each other. In this way a library was built that totaled approximately 59,000 sgRNA sequences, where each sgRNA by itself is able to simultaneously modify two to 10 genes at once from each gene family, thereby effectively neutralizing the phenomenon of genetic redundancy.
In addition, the sgRNA sequences were divided into ten sub-libraries of approximately 6,000 sgRNA sequences each, according to the presumed role of the genes—such as coding for enzymes, receptors, transcription factors, etc. According to the researchers, establishing the libraries allowed them to focus and optimize the search for genes responsible for desired traits, a search that until now has been largely random.
In the next step, the researchers moved from the computer to the laboratory. Here, they generated all 59,000 sgRNA sequences designed by the computational method and engineered them into new plasmid libraries (i.e., circular DNA segments) in combination with the cutting enzyme. The researchers then generated thousands of new plants containing the libraries—where each plant was implanted with a single sgRNA sequence directed against a specific gene family.
The researchers observed the traits that were manifested in the plants following the genome modifications, and when an interesting phenotype was observed in a particular plant. it was easy to know which genes were responsible for the change based on the sgRNA sequence that was inserted into it.
Also, through DNA sequencing of the identified genes, it was possible to determine the nature of the mutation that caused the change and its contribution to the plant's new properties. In this way, many new traits were mapped that until now were blocked due to genetic redundancy. Specifically, the researchers identified specific proteins that comprise a mechanism related to the transport of the hormone cytokinin, which is essential for optimal plant development.
Prof. Shani says, "The new method we developed is expected to be of great help to basic research in understanding processes in plants, but beyond that, it has enormous significance for agriculture: it makes it possible to efficiently and accurately reveal the pool of genes responsible for traits we seek to improve—such as resistance to drought, pests, and diseases, or increasing yields. We believe that this is the future of agriculture: controlled and targeted crop improvement on a large scale. Today we are applying the method we developed to rice and tomato plants with great success, and we intend to apply it to other crops as well."
More information: Yangjie Hu et al, Multi-Knock—a multi-targeted genome-scale CRISPR toolbox to overcome functional redundancy in plants, Nature Plants (2023). DOI: 10.1038/s41477-023-01374-4
Journal information: Nature Plants
Provided by Tel Aviv University